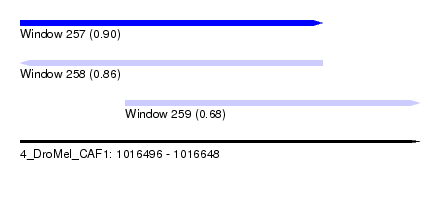
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,016,496 – 1,016,648 |
| Length | 152 |
| Max. P | 0.902857 |

| Location | 1,016,496 – 1,016,611 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -27.67 |
| Consensus MFE | -25.46 |
| Energy contribution | -25.13 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902857 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1016496 115 + 1281640 AAGAAGUAGGAAAUACUUUUGGCCUUCAGUAAUUUAAAACG-CUCUAAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUACUGUUCGAAGGCUCGUACGCACAUCC .(((((((.....)))))))((((((((((((((((.....-...))))).....))))((((((.(((((((.......)))))))))))))...)))))))............. ( -28.30) >DroSec_CAF1 5035 116 + 1 AAGAAGUAGGAAAUACUUUUGGUCUUCAGUAAUUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCC .(((((((.....)))))))((((((((....)....((((((...((((......))))......(((((((.......)))))))..)))))).)))))))............. ( -27.60) >DroSim_CAF1 4954 116 + 1 AAGAAGUAGGAAAUACUUUUGGUCUUCAGUAACUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCC .(((((((.....)))))))(((((((..........((((((...((((......))))......(((((((.......)))))))..)))))).)))))))............. ( -27.10) >consensus AAGAAGUAGGAAAUACUUUUGGUCUUCAGUAAUUUAAAAUGGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCC .(((((((.....)))))))(((((((...................((((......)))).((((.(((((((.......))))))))))).....)))))))............. (-25.46 = -25.13 + -0.33)

| Location | 1,016,496 – 1,016,611 |
|---|---|
| Length | 115 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 93.10 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -24.12 |
| Energy contribution | -23.57 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.856986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1016496 115 - 1281640 GGAUGUGCGUACGAGCCUUCGAACAGUAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUUAGAG-CGUUUUAAAUUACUGAAGGCCAAAAGUAUUUCCUACUUCUU (((.((((....(.((((((...((((((..(((((.......)))))...))))))((((....(((......-.)))......)))))))))))....)))).)))........ ( -30.10) >DroSec_CAF1 5035 116 - 1 GGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAAUUACUGAAGACCAAAAGUAUUUCCUACUUCUU (((.((((((....(((((((((.(((....(((((.......)))))........((((......))))...))).))).........))))))...)))))).)))........ ( -26.30) >DroSim_CAF1 4954 116 - 1 GGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAGUUACUGAAGACCAAAAGUAUUUCCUACUUCUU (((.((((((....((((((.((((((....(((((.......)))))........((((......))))...))).......)))...))))))...)))))).)))........ ( -26.71) >consensus GGAUGUGCUUACAAGUCUUCGAACGGCAUUAUGUACAUAAAUAGUACACAUAUACUGAGUAUGAUGACUCUAAGCCAUUUUAAAUUACUGAAGACCAAAAGUAUUUCCUACUUCUU (((.((((((....((((((...(((.((..(((((.......)))))...)).)))((((.((((.(.....).))))......))))))))))...)))))).)))........ (-24.12 = -23.57 + -0.55)

| Location | 1,016,536 – 1,016,648 |
|---|---|
| Length | 112 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 93.57 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -24.84 |
| Energy contribution | -26.40 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1016536 112 + 1281640 G-CUCUAAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUACUGUUCGAAGGCUCGUACGCACAUCCUU-UUAGGCCUUGCAUUCGCAAGCGUCAGUAAAUAAGA (-(....((((.((.....((((((.(((((((.......)))))))))))))...)).)))).....))........-...((((((((....))))).)))........... ( -28.30) >DroSec_CAF1 5075 114 + 1 GGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUAGGCCUUGCAUUCGCUAGCGUCAGUAAAUAAGA .((((((((((.((..((...((((.(((((((.......))))))))))).))..)).))))).)))))............(((((.((....)).)).)))........... ( -27.20) >DroSim_CAF1 4994 114 + 1 GGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUAGGCCUUGCAUUCGCAAGCGUCAGUAAAUAAGA .((((((((((.((..((...((((.(((((((.......))))))))))).))..)).))))).)))))............((((((((....))))).)))........... ( -31.30) >consensus GGCUUAGAGUCAUCAUACUCAGUAUAUGUGUACUAUUUAUGUACAUAAUGCCGUUCGAAGACUUGUAAGCACAUCCUUUUUAGGCCUUGCAUUCGCAAGCGUCAGUAAAUAAGA .((((((((((.((..((...((((.(((((((.......))))))))))).))..)).))))).)))))............((((((((....))))).)))........... (-24.84 = -26.40 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:41 2006