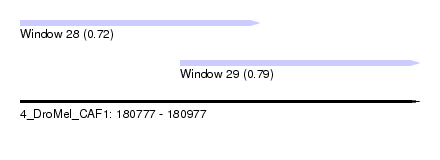
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 180,777 – 180,977 |
| Length | 200 |
| Max. P | 0.788774 |

| Location | 180,777 – 180,897 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.75 |
| Mean single sequence MFE | -31.66 |
| Consensus MFE | -23.56 |
| Energy contribution | -23.76 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723946 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 180777 120 + 1281640 UAGGAGGUAGGUGACCAUUAAAGUGUCAUUCGUUUUAGCAAAAACACUUUGUAGUUGCACACCGUGAAAUAUAUCCCUGUAAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAA ..((((((..(..((...((((((((..((.((....))))..))))))))..))..).(.((.(((..((((....)))).)))((((((((....)))))))))).)))))))..... ( -31.90) >DroSec_CAF1 7109 120 + 1 UAGGAGGUAGGUGACCAUUAAAGUGUCAUUCGUCUUAGCAAACACACUUUGUAGUUUCACACCGUGAAAUAUAUCCCUAUAAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAA ..((((((.(((((((......).))))))((((...(((((.....))))).(((((((...)))))))...............((((((((....))))))))))))))))))..... ( -32.80) >DroSim_CAF1 4896 120 + 1 UAGGAGGCAGGUGACCAUUAAAGUGUCAUUCGUCCCAGCAAACACACUUUGUAGUUUCCCACCGUGAACUAUAUCCCUAUAAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAA ..((((((.(((((((......).))))))((((...............(((((((..(....)..)))))))............((((((((....))))))))))))))))))..... ( -34.00) >DroEre_CAF1 9653 120 + 1 UAGGAGGUAGGUGACCAUUAACGUGUCAUUCGUUUUAGUCAAAGCAGUUAGUAGUUGCACACCGUGAAAUAUAUCCCUAUAAUCAGAUUGCAUAACCAUGCGAUCAGCGGCUUCUAACAA ...((((..(((((((......).))))))..))))..........(((((.((((((......(((..((((....)))).)))((((((((....)))))))).)))))).))))).. ( -30.70) >DroYak_CAF1 6859 120 + 1 AAGGAGGUAGGUGAUCAAUAACGUUUUCUUGGUUUGAGUCAAAGCUGUUAGUAGUUGCACACCGUGAAAUAUAUCUCUAUAAUCAGAUUGCAUUACCAUGCGAUCGGCGGCUUCCAACAA ..((((((.((((..((((.((.......((((((......))))))...)).))))..))))......................((((((((....))))))))....))))))..... ( -28.90) >consensus UAGGAGGUAGGUGACCAUUAAAGUGUCAUUCGUUUUAGCAAAAACACUUUGUAGUUGCACACCGUGAAAUAUAUCCCUAUAAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAA ..((((((.(((((((......).))))))((((...((((..((.....))..))))...........................((((((((....))))))))))))))))))..... (-23.56 = -23.76 + 0.20)



| Location | 180,857 – 180,977 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.92 |
| Mean single sequence MFE | -25.73 |
| Consensus MFE | -19.76 |
| Energy contribution | -19.92 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 180857 120 + 1281640 AAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAAUAUUUUACUAAUAUGUCGAUAUUCUAUUAAGGAAUUACUUUUUAUGUUAUUGCUGAAUUUUAAUAAAUGGUUUUCCGUCC .....((((((((....))))))))(((((...(((...(((((.....)))))(.(((((...((..(((......)))..))...))))))..............)))....))))). ( -23.40) >DroSec_CAF1 7189 120 + 1 AAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAAUAUUUUACUAAUAUGUCGAUAUAAGGAUAAGGAAUUUCUUUUUGUUUUAUUGCUGAAUUUUAAUAAAUGGUUUUCCGCCC .....((((((((....))))))))(((((...(((...(((((.....)))))(.(((((.((.((.((((....)))).)).)).))))))..............)))....))))). ( -27.10) >DroSim_CAF1 4976 120 + 1 AAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAAUAUUUUACUAGUAUGUCGAUAUAAGGAUAAGGAAUUACUUUUUGUAUUAUUGCUGAAUUUUAAUAAAUGGUUUUCCGCCC .....((((((((....))))))))(((((...(((............(((((....(((((((((((((....))).))))))))))..)))))............)))....))))). ( -28.75) >DroEre_CAF1 9733 120 + 1 AAUCAGAUUGCAUAACCAUGCGAUCAGCGGCUUCUAACAAUAUUUUACUAUUAUGUCGAUAUUCGAUUGAGGAAUUACUUUUGUUUUUAUUGCUGAGAUUUAAUAGGUAACUUUCCGCCC .....((((((((....))))))))...(((.......................((((.....))))...((((((((((..(((((.......))))).....))))))..))))))). ( -24.20) >DroYak_CAF1 6939 120 + 1 AAUCAGAUUGCAUUACCAUGCGAUCGGCGGCUUCCAACAAUAUUUUACUAAUAUUUCGAUAUUCGAUUGAAAAAUUACUUUUUCUUUUAUUGCUGAAUUUUAAUAAGUGACUUUCCGCCC .....((((((((....))))))))(((((..................((((.(((((((.....))))))).)))).....((.(((((((........))))))).))....))))). ( -25.20) >consensus AAUCAGAUUGCAUAACCAUGCGAUCGGCGGCUUCCAAUAAUAUUUUACUAAUAUGUCGAUAUUCGAUUAAGGAAUUACUUUUUGUUUUAUUGCUGAAUUUUAAUAAAUGGUUUUCCGCCC .....((((((((....))))))))(((((...(((...(((((.....)))))..(((((.......(((......))).......)))))...............)))....))))). (-19.76 = -19.92 + 0.16)

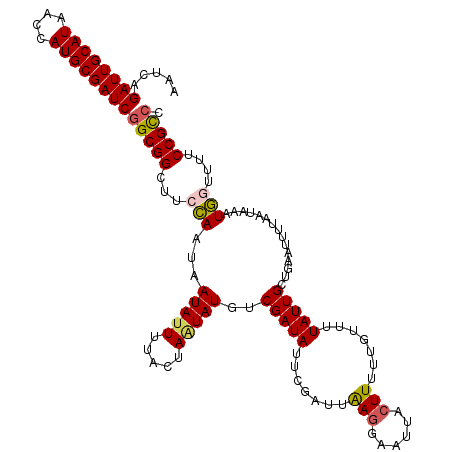

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:26 2006