| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,016,381 – 1,016,472 |
| Length | 91 |
| Max. P | 0.931512 |

| Location | 1,016,381 – 1,016,472 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -17.14 |
| Energy contribution | -17.08 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931512 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
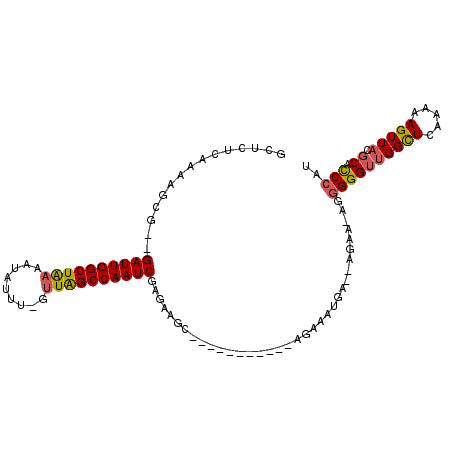
>4_DroMel_CAF1 1016381 91 + 1281640 GCUUUCAAAAUCG--GAUUGGCUAAAAUAUUU-GUUAGCCAAUCGAGAAGC-----------AGAAAGGA--AGAA-UGGGGGUUGGCUCAAAAAGUUACGCACCCAU .(((((....((.--((((((((((.......-.))))))))))..))...-----------.)))))..--...(-((((.(((((((.....))))).)).))))) ( -27.80) >DroSec_CAF1 4920 91 + 1 GCUCUCAAAAUCG--GAUUGGCUAAAAUAUUU-GUUAGCCAAUCGAGAAGC-----------AGAAAUGA--AGAA-UGGGGGUUGGCUCAAAAAGUUACGCACCCAU ((((((.......--((((((((((.......-.))))))))))))).)))-----------........--...(-((((.(((((((.....))))).)).))))) ( -27.11) >DroSim_CAF1 4839 91 + 1 GCUCUCAAAAUCG--GAUUGGCUAAAAUAUUU-GUUAGCCAAUCGAGAAGC-----------AGAAAUGA--AGAA-UGGGGGUUGGCUCAAAAAGUUACGCACCCAU ((((((.......--((((((((((.......-.))))))))))))).)))-----------........--...(-((((.(((((((.....))))).)).))))) ( -27.11) >DroEre_CAF1 4378 92 + 1 GCUCUCAAAAGCG--GAUUGGCUGAAAUAUUUGGUUAGCCAAUCGAGAAGC-----------AGCAAUAA--AGCA-AGGGGGUUGGCUCAAAAAGUUACGCAUCCAU ((((((.......--((((((((((.........))))))))))))).)))-----------.((.....--.)).-.((..(((((((.....))))).))..)).. ( -24.41) >DroYak_CAF1 6224 92 + 1 GCUCUCAAAAGCG--GAUUGGCUGAAAUAUUU-GUUGGCCAAUCAAGAAGC-----------AGAAAUAA--AGCAAAGGGGGUUGGCUCGAAAAGUUACGCACCCAU (((.......((.--(((((((..(.......-.)..))))))).....))-----------........--)))...(((.(((((((.....))))).)).))).. ( -24.16) >DroPer_CAF1 39771 103 + 1 GCUCUCGAACGCGGGGAUUGGCUA---UCGUC-GUUAGCCAAUCGUGUCUCGUCGUUACCGUUGCAAUGUUUCGUU-AGAGGGUAGGUUACGAAAGUUACUCUUCAAU ((((((.(((((((((((((((((---.....-..)))))))))....)))).))))..((..((...))..))..-.)))))).(((.((....)).)))....... ( -32.00) >consensus GCUCUCAAAAGCG__GAUUGGCUAAAAUAUUU_GUUAGCCAAUCGAGAAGC___________AGAAAUGA__AGAA_AGGGGGUUGGCUCAAAAAGUUACGCACCCAU ...............((((((((((.........))))))))))...................................((((((((((.....))))).)).))).. (-17.14 = -17.08 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:37 2006