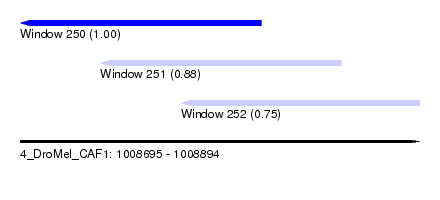
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 1,008,695 – 1,008,894 |
| Length | 199 |
| Max. P | 0.999925 |

| Location | 1,008,695 – 1,008,815 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -34.38 |
| Energy contribution | -34.42 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
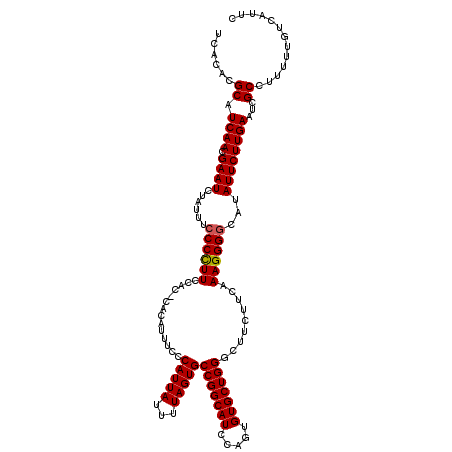
>4_DroMel_CAF1 1008695 120 - 1281640 CUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAAUUUCAUGGCGCAUUGUAAAGGUGACGAAUCUGAUUUUCCUGGAAUGUGUGAAAAUACCUUA .....((((.(((((((((.((((((....(((((((..(((.((((((((((((.........))))))).)))))))))))))))..))))))....))))))))).......)))). ( -37.10) >DroSec_CAF1 48355 120 - 1 CUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAAUUUCAUGGCGCAUUGUAAAGGUGACGAAUCUGAUUUUCCUGGAAUGUGUGAAAAUACCAUA ..........(((((((((.((((((....(((((((..(((.((((((((((((.........))))))).)))))))))))))))..))))))....)))))))))............ ( -35.80) >DroSim_CAF1 33917 120 - 1 CUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAAUUUCAUGGCGCAUUGUAAAGGUGACGAAUCUGAUUUUCCUGGAAUGUGUGAAAAUACCAUA ..........(((((((((.((((((....(((((((..(((.((((((((((((.........))))))).)))))))))))))))..))))))....)))))))))............ ( -35.80) >DroEre_CAF1 50609 120 - 1 CUUCAAAGGGAAACAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAACUUCAUGGCGCAUUGUAAAGGUGACGAAUCUGAUUUUCCUGGAAUGUGUGAAAAUACCUUA .....((((...(((((((.((((((....(((((((..(((.((((((((((((.........))))))).)))))))))))))))..))))))....)))))))((....)).)))). ( -36.10) >DroYak_CAF1 48992 120 - 1 CUUCAAAGGGGAACAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAACUUCAUGGCGCAUUGUAGAGGUGACGAAUCAGAUUUUCCUGGAAUGUGUGAAAAUACCUUA .....((((...(((((((.(((((.....((((((((..((((...((((((((.........)))))))).))))..))))))))...)))))....)))))))((....)).)))). ( -38.80) >consensus CUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAAUUUCAUGGCGCAUUGUAAAGGUGACGAAUCUGAUUUUCCUGGAAUGUGUGAAAAUACCUUA ..........(((((((((.((((((....(((((((..(((.((((((((((((.........))))))).)))))))))))))))..))))))....)))))))))............ (-34.38 = -34.42 + 0.04)


| Location | 1,008,735 – 1,008,855 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -34.15 |
| Consensus MFE | -30.48 |
| Energy contribution | -30.92 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.876917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1008735 120 - 1281640 UUUCCAUUAUUGUAGUGCCGGCAUCCAGUGUGCUGGGCUUCUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAAUUUCAUGGCGCAUUGUAAAGG ...........(.(((.(((((((.....)))))))))).)..(((((((((.............))))))))).....(((.((((((((((((.........))))))).)))))))) ( -34.42) >DroSec_CAF1 48395 120 - 1 UUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGCUUCUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAAUUUCAUGGCGCAUUGUAAAGG .............(((.(((((((.....))))))))))....(((((((((.............))))))))).....(((.((((((((((((.........))))))).)))))))) ( -34.12) >DroSim_CAF1 33957 120 - 1 UUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGCUUCUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAAUUUCAUGGCGCAUUGUAAAGG .............(((.(((((((.....))))))))))....(((((((((.............))))))))).....(((.((((((((((((.........))))))).)))))))) ( -34.12) >DroEre_CAF1 50649 120 - 1 UUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGGUUCUUCAAAGGGAAACAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAACUUCAUGGCGCAUUGUAAAGG ...((.((((..(.((((((((..((((....))))(((((.......(....)......))))))))((((((.(((.((....).).))).))))))......))))))..)))).)) ( -32.32) >DroYak_CAF1 49032 120 - 1 UUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGGCUCUUCAAAGGGGAACAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAACUUCAUGGCGCAUUGUAGAGG (((((........(((.(((((((.....))))))).)))........)))))..........................(((.((((((((((((.........))))))).)))))))) ( -35.79) >consensus UUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGCUUCUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUCCUAUUGCGUGUGCCAAAAAUUUCAUGGCGCAUUGUAAAGG .................(((((((.....))))))).......(((((((((.............))))))))).....(((.((((((((((((.........))))))).)))))))) (-30.48 = -30.92 + 0.44)

| Location | 1,008,775 – 1,008,894 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.65 |
| Mean single sequence MFE | -25.36 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.14 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.748535 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 1008775 119 - 1281640 UCACACGCAUCAACGAAUCUAUUUCCCCUUCCAC-CACAAUUUCCAUUAUUGUAGUGCCGGCAUCCAGUGUGCUGGGCUUCUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUC ......((.((((.((((......((((((.(((-.(((((.......))))).)))(((((((.....))))))).........))))))...))))))))...))............. ( -27.50) >DroSec_CAF1 48435 119 - 1 UCACACGCAUCAACGAAUCUAUUUCCCCUUCCAC-CACAUUUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGCUUCUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUC ......((.((((.((((......((((((....-.........(((((...)))))(((((((.....))))))).........))))))...))))))))...))............. ( -22.30) >DroSim_CAF1 33997 119 - 1 UCACACGCAUCAACGAAUCUAUUUCCCCUUCCAC-CACAUUUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGCUUCUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUC ......((.((((.((((......((((((....-.........(((((...)))))(((((((.....))))))).........))))))...))))))))...))............. ( -22.30) >DroEre_CAF1 50689 120 - 1 UCACACGCAUCAACGAAUCUAUUUCCCUUUCCCCUCGCAUUUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGGUUCUUCAAAGGGAAACAUUCUUGAAUCGCCUUUUUGUCAUUC ......((.((((.((((...(((((((((((((..(((((............))))).(((((.....)))))))))......))))))))).))))))))...))............. ( -28.00) >DroYak_CAF1 49072 120 - 1 UCACACGCAUCAACGAAUCUAUUCCCCCUUCCCCUCGCAUUUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGGCUCUUCAAAGGGGAACAUUCUUGAAUCGCCUUUUUGUCAUUC ......((.((((.((((...((((((.........(((((............))))).(((.(((((....)))))))).......)))))).))))))))...))............. ( -26.70) >consensus UCACACGCAUCAACGAAUCUAUUUCCCCUUCCAC_CACAUUUCCCAUUAUUUUAGUGCCGGCAUCCAGUGUGCUGGGCUUCUUCAAAGGGGCAUAUUCUUGAAUCGCCUUUUUGUCAUUC ......((.((((.((((......((((((..............(((((...)))))(((((((.....))))))).........))))))...))))))))...))............. (-21.10 = -21.14 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:32 2006