| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 179,842 – 180,039 |
| Length | 197 |
| Max. P | 0.905913 |

| Location | 179,842 – 179,962 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.92 |
| Mean single sequence MFE | -17.96 |
| Consensus MFE | -13.50 |
| Energy contribution | -12.58 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 179842 120 - 1281640 GAAUAUAAUGAAGUGAAUAAUCAUUUCAUAUCACCAGAAACACACAAAAAAACAAUUGUGAUUGAAAAUUACACGAACGCAAUAGCUUCCACAAAAGUACAAGACUUAAAUAUAAAAGGA .......(((((((((....)))))))))....((.......(((((........)))))..............(((.((....))))).....((((.....))))..........)). ( -16.30) >DroSec_CAF1 6164 120 - 1 GAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAACACACAAAAAAACAAUUGUGGUUCAAAAUUACACGAACGCAAUAGCUUCAACAAAAGUAGAAGAAUUAAAUAUAAAAGGA .......(((((((((....)))))))))....((.(((...(((((........))))).))).....................((((.((....)).))))..............)). ( -20.10) >DroSim_CAF1 3951 120 - 1 GAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAACACACAAAAAAACAAUUGUGGUUCAAAAUUACACGAACGCAAUAGCUUCCACAAAAGUAGAAGACUUAAAUAUAAAAGGA .......(((((((((....)))))))))....((.(((...(((((........))))).)))...................((((((.((....)).)))).))...........)). ( -20.30) >DroEre_CAF1 8709 120 - 1 AAAUAUAAUGGAGUGAAGAAUCAUUUUAUGAUACCAGGAUCACAUACAAAAAGAAGUAUAAUUCAAACUUACACGAACGCGAUAGCUUCUAGAAAAGUACAUGGUUUAAAUAUCACACGU .......(((((((((....)))))))))((((...((((((..(((.....(((......)))..........(((.((....))))).......)))..))))))...))))...... ( -18.00) >DroYak_CAF1 5909 120 - 1 AAAUAUAAUGGAGUGAAGAAUCAUUUCAUGAAUACAGAAACGCAUACAAAAACAAGUAUGAUUCAAACUUACACGAACACGAUAGCUUCUAGAAAAGUACACGGCUUAAAUAUCACACGU .......(((((((((....)))))))))((.....(((...(((((........))))).)))...............((...((((......))))...)).........))...... ( -15.10) >consensus GAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAACACACAAAAAAACAAUUGUGAUUCAAAAUUACACGAACGCAAUAGCUUCCACAAAAGUACAAGACUUAAAUAUAAAAGGA .......(((((((((....))))))))).......(((...(((((........))))).)))..........(((.((....)))))............................... (-13.50 = -12.58 + -0.92)


| Location | 179,882 – 179,999 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.03 |
| Mean single sequence MFE | -20.54 |
| Consensus MFE | -14.50 |
| Energy contribution | -13.26 |
| Covariance contribution | -1.24 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 179882 117 - 1281640 AUAGAAUUUGAAUAG---UAAUUCUAGUUUUACCUUACAGGAAUAUAAUGAAGUGAAUAAUCAUUUCAUAUCACCAGAAACACACAAAAAAACAAUUGUGAUUGAAAAUUACACGAACGC .(((((((.......---.)))))))(((((.((.....))......(((((((((....))))))))).......))))).(((((........))))).................... ( -20.40) >DroSec_CAF1 6204 117 - 1 AUAGGAUUUGAAUAA---UAAUUCCAGUUUUACCUUACAGGAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAACACACAAAAAAACAAUUGUGGUUCAAAAUUACACGAACGC ......(((((((..---..(((((.((........)).)))))...(((((((((....))))))))).............(((((........))))))))))))............. ( -21.30) >DroSim_CAF1 3991 117 - 1 AUAGGAUUUGAAUAA---UAAUUCCAGUUUUACCUUACAGGAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAACACACAAAAAAACAAUUGUGGUUCAAAAUUACACGAACGC ......(((((((..---..(((((.((........)).)))))...(((((((((....))))))))).............(((((........))))))))))))............. ( -21.30) >DroEre_CAF1 8749 117 - 1 AAAGAAUUCAAAUAA---UAAUUUUGGUUUUACCUUAUAGAAAUAUAAUGGAGUGAAGAAUCAUUUUAUGAUACCAGGAUCACAUACAAAAAGAAGUAUAAUUCAAACUUACACGAACGC ...(((((.......---..((((((((.(((..(((((....)))))((((((((....))))))))))).))))))))...((((........)))))))))................ ( -17.60) >DroYak_CAF1 5949 120 - 1 AUAGAGUUCAAAUAAUAAUAAUUCUGGAUUUACCUUACAGAAAUAUAAUGGAGUGAAGAAUCAUUUCAUGAAUACAGAAACGCAUACAAAAACAAGUAUGAUUCAAACUUACACGAACAC .....((((........(((.(((((...........))))).))).(((((((((....))))))))).......(((...(((((........))))).)))..........)))).. ( -22.10) >consensus AUAGAAUUUGAAUAA___UAAUUCUAGUUUUACCUUACAGGAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAACACACAAAAAAACAAUUGUGAUUCAAAAUUACACGAACGC .(((((((.((((.......)))).)))))))...............(((((((((....))))))))).......(((...(((((........))))).)))................ (-14.50 = -13.26 + -1.24)

| Location | 179,922 – 180,039 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.20 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -15.28 |
| Energy contribution | -14.78 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 179922 117 - 1281640 UCAUGACAUUAAUAUCUAAGAAGGUAACUGUUAAACUCUCAUAGAAUUUGAAUAG---UAAUUCUAGUUUUACCUUACAGGAAUAUAAUGAAGUGAAUAAUCAUUUCAUAUCACCAGAAA ......................(((..((((..........(((((((.......---.)))))))..........)))).......(((((((((....)))))))))...)))..... ( -20.75) >DroSec_CAF1 6244 117 - 1 UCAUGACAUUAAUAUCUAAGAAGGUAACUGUUAAACUCUCAUAGGAUUUGAAUAA---UAAUUCCAGUUUUACCUUACAGGAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAA ..............(((...(((((((((((((((..(.....)..)))))....---......))))..))))))...))).....(((((((((....)))))))))........... ( -19.10) >DroSim_CAF1 4031 117 - 1 UCAUGACAUUAAUAUCUAAGAAGGUAAUUGUUAAACUCUCAUAGGAUUUGAAUAA---UAAUUCCAGUUUUACCUUACAGGAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAA ...(((((....(((((....)))))..)))))..........((..........---..(((((.((........)).)))))...(((((((((....)))))))))....))..... ( -19.60) >DroYak_CAF1 5989 120 - 1 ACAUGAUAUUAAUGUCUAAUAAGGUAAGUUUAAGAGUCUUAUAGAGUUCAAAUAAUAAUAAUUCUGGAUUUACCUUACAGAAAUAUAAUGGAGUGAAGAAUCAUUUCAUGAAUACAGAAA ...((.((((....(((..(((((((((((((.((((.((((............))))..))))))))))))))))).)))......(((((((((....))))))))).)))))).... ( -21.90) >consensus UCAUGACAUUAAUAUCUAAGAAGGUAACUGUUAAACUCUCAUAGAAUUUGAAUAA___UAAUUCCAGUUUUACCUUACAGGAAUAUAAUGAAGUGAAUAAUCAUUUCAUAACACCAGAAA ..............(((...(((((((..............(((((((...........)))))))...)))))))..)))......(((((((((....)))))))))........... (-15.28 = -14.78 + -0.50)

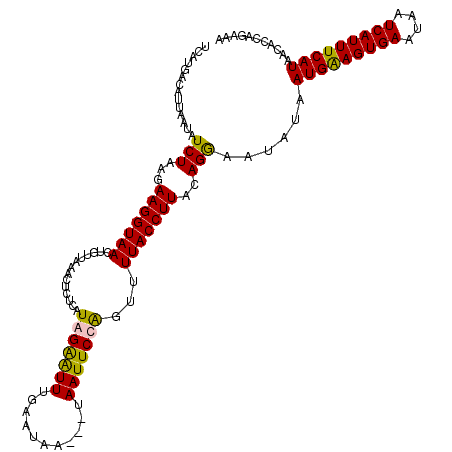
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:22 2006