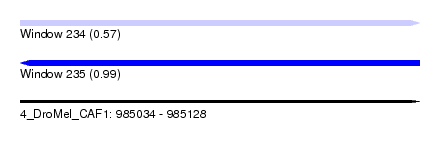
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 985,034 – 985,128 |
| Length | 94 |
| Max. P | 0.991119 |

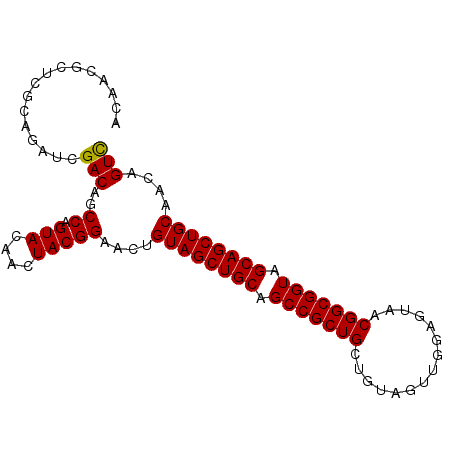
| Location | 985,034 – 985,128 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -25.20 |
| Energy contribution | -25.08 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 985034 94 + 1281640 GACUGUUGCAGCUGCUACCGCCGUUACUCCAACUACAGCAGCGGCUGCAGCUACUGUUCCGUAGUUGUACUGGCUGUCGAUCUGCGAGCGUUGU (((.((((((((((((...((.((..........)).))))))))))))))....((((.(((((((.((.....))))).))))))))))).. ( -32.50) >DroSec_CAF1 18017 94 + 1 GACUGUUGCAGCUGCUACCGCCGUUACACCAACCACAGCAGCGGCUGCAGCUACAGUUCCGUAGUUGUACUGGCUGUCGAUCUGCGAGCGUUGU (((.((((((((((((...((.((..........)).))))))))))))))....((((.(((((((.((.....))))).))))))))))).. ( -32.00) >DroSim_CAF1 15661 94 + 1 GACUGUUGCAGCUGCUACCGCCGUUACACCAACUACAGCAGCGGCUGCAGCUACAGUUCCGUAGUUGUACUGGCUGUCGAUCUGCGAGCGUUGU (((.((((((((((((...((.((..........)).))))))))))))))....((((.(((((((.((.....))))).))))))))))).. ( -32.70) >DroEre_CAF1 18864 91 + 1 GACAGUUGCAGCUGCUACCGCCGUUACUCCAACUACAGCAGCGGCUGCAGCUACAGUACCGUAGUUGUACUGGUUGUCUAUCUG---CCGUUGU ((((((((((((((((...((.((..........)).))))))))))))))).((((((.......))))))...)))......---....... ( -33.10) >DroYak_CAF1 19209 91 + 1 AACAGUUGCAGCUGCUACCGCCGUCACUCCAACCACUGCAGCGGCUGCAGCUACAGUACCGUAAUUGUACUGGCUGUCUAUCGG---GCGUUGU ((((((((((((((((...((.((..........)).))))))))))))))).((((((.......))))))...(((.....)---))))).. ( -31.70) >consensus GACUGUUGCAGCUGCUACCGCCGUUACUCCAACUACAGCAGCGGCUGCAGCUACAGUUCCGUAGUUGUACUGGCUGUCGAUCUGCGAGCGUUGU (((.((((((((((((...((.((..........)).)))))))))))))).(((((...(((....)))..)))))............))).. (-25.20 = -25.08 + -0.12)

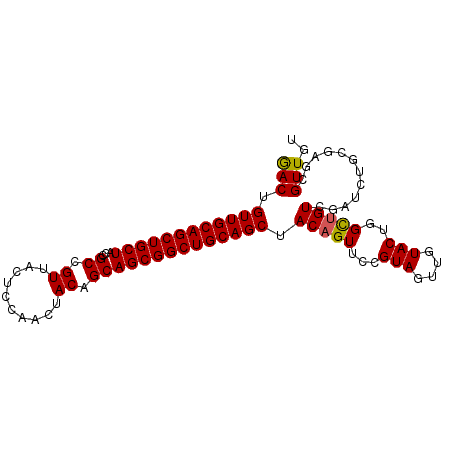
| Location | 985,034 – 985,128 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 91.46 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -29.60 |
| Energy contribution | -29.44 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 985034 94 - 1281640 ACAACGCUCGCAGAUCGACAGCCAGUACAACUACGGAACAGUAGCUGCAGCCGCUGCUGUAGUUGGAGUAACGGCGGUAGCAGCUGCAACAGUC .........((((....((.(((.((.(((((((((..(((..((....))..)))))))))))).....))))).)).....))))....... ( -31.00) >DroSec_CAF1 18017 94 - 1 ACAACGCUCGCAGAUCGACAGCCAGUACAACUACGGAACUGUAGCUGCAGCCGCUGCUGUGGUUGGUGUAACGGCGGUAGCAGCUGCAACAGUC .....(((.((((.(((.....).(((....))).)).)))))))((((((.((((((((.((((...)))).)))))))).))))))...... ( -34.20) >DroSim_CAF1 15661 94 - 1 ACAACGCUCGCAGAUCGACAGCCAGUACAACUACGGAACUGUAGCUGCAGCCGCUGCUGUAGUUGGUGUAACGGCGGUAGCAGCUGCAACAGUC .....(((.((((.(((.....).(((....))).)).)))))))((((((.((((((((.((((...)))).)))))))).))))))...... ( -32.10) >DroEre_CAF1 18864 91 - 1 ACAACGG---CAGAUAGACAACCAGUACAACUACGGUACUGUAGCUGCAGCCGCUGCUGUAGUUGGAGUAACGGCGGUAGCAGCUGCAACUGUC .....((---(((.........((((((.......))))))....((((((.((((((((.((((...)))).)))))))).)))))).))))) ( -36.70) >DroYak_CAF1 19209 91 - 1 ACAACGC---CCGAUAGACAGCCAGUACAAUUACGGUACUGUAGCUGCAGCCGCUGCAGUGGUUGGAGUGACGGCGGUAGCAGCUGCAACUGUU .......---......(((((..(((((.......)))))((((((((.(((((((((.(......).)).))))))).))))))))..))))) ( -37.30) >consensus ACAACGCUCGCAGAUCGACAGCCAGUACAACUACGGAACUGUAGCUGCAGCCGCUGCUGUAGUUGGAGUAACGGCGGUAGCAGCUGCAACAGUC ................(((..((.(((....)))))....((((((((.(((((((...............))))))).))))))))....))) (-29.60 = -29.44 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:37:02 2006