
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 957,450 – 957,559 |
| Length | 109 |
| Max. P | 0.992336 |

| Location | 957,450 – 957,559 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -23.86 |
| Energy contribution | -24.43 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.32 |
| SVM RNA-class probability | 0.992336 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 957450 109 + 1281640 UU-UU-AGGGCUUUCGCUAACACAUUGCUUUUGCAGCUUCAGAGCAACGCCAUCAUCAGUACCAUUCUUUUUGGUAC---UUGGCUUGUCCUUACAUUCCAAAGCUUUUGCUGU ..-..-(((((....(((......((((....))))......)))...((((.....(((((((.......))))))---)))))..)))))..........(((....))).. ( -28.90) >DroSim_CAF1 17793 112 + 1 UU-UU-AGGGCUUUCGCUAACACAUUGCUUUUGCAGCUUCAGAGCAACGCCAUCACCAGUACCAUUCUUUUUGGUACUUCUUGGCGUGUCCUUACAUUCCAAAGCUUUUUCUGU ..-..-(((((....(((......((((....))))......))).((((((.....(((((((.......)))))))...)))))))))))...................... ( -31.10) >DroEre_CAF1 23177 107 + 1 UUUUUAAGGGCUUUUGCUAACACAUUGCUUUUGCAGCUUCAGAGCAACGCCAUCAUCAGUACCAUUCUUUUUGGUACUUCUUUGCUUGUCCUUACAUUUCGAAGCUU------- ....(((((((..(((((......((((....))))......))))).((.......(((((((.......))))))).....))..))))))).............------- ( -25.50) >DroYak_CAF1 22900 113 + 1 UUUUU-AGGGCUUUUGCUAACACAUUGCUUUUGCAGCUUCAGAGCAACGCCAUCAUCAGUACCAUCCUAUUUGGUACUUCUUGUCUUGCCCUUACAUUCCAAAGCUUUUGCUGU .....-(((((..(((((......((((....))))......)))))..........(((((((.......))))))).........)))))..........(((....))).. ( -27.40) >consensus UU_UU_AGGGCUUUCGCUAACACAUUGCUUUUGCAGCUUCAGAGCAACGCCAUCAUCAGUACCAUUCUUUUUGGUACUUCUUGGCUUGUCCUUACAUUCCAAAGCUUUUGCUGU ......(((((....(((......((((....))))......)))...((((.....(((((((.......)))))))...))))..)))))...................... (-23.86 = -24.43 + 0.56)

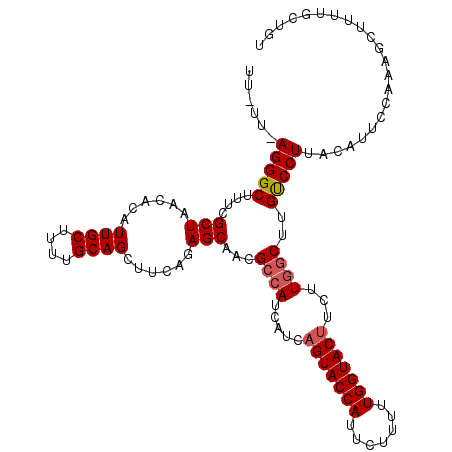
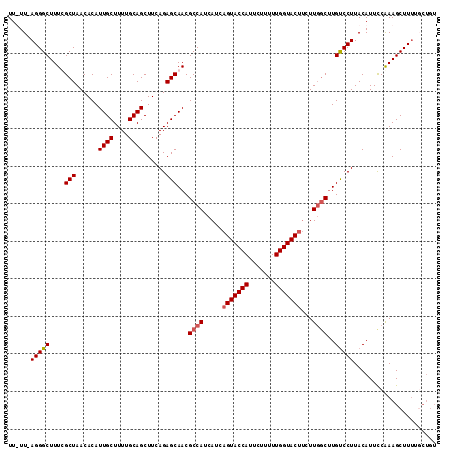
| Location | 957,450 – 957,559 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -23.51 |
| Energy contribution | -23.82 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 957450 109 - 1281640 ACAGCAAAAGCUUUGGAAUGUAAGGACAAGCCAA---GUACCAAAAAGAAUGGUACUGAUGAUGGCGUUGCUCUGAAGCUGCAAAAGCAAUGUGUUAGCGAAAGCCCU-AA-AA ..((((..(((((..((.(((....))).(((((---((((((.......))))))).....)))).....))..)))))((....))....)))).((....))...-..-.. ( -30.50) >DroSim_CAF1 17793 112 - 1 ACAGAAAAAGCUUUGGAAUGUAAGGACACGCCAAGAAGUACCAAAAAGAAUGGUACUGGUGAUGGCGUUGCUCUGAAGCUGCAAAAGCAAUGUGUUAGCGAAAGCCCU-AA-AA (((.....(((((..((..((((.(.(((((((....((((((.......))))))))))).)).).))))))..)))))((....))..)))....((....))...-..-.. ( -33.30) >DroEre_CAF1 23177 107 - 1 -------AAGCUUCGAAAUGUAAGGACAAGCAAAGAAGUACCAAAAAGAAUGGUACUGAUGAUGGCGUUGCUCUGAAGCUGCAAAAGCAAUGUGUUAGCAAAAGCCCUUAAAAA -------.(((((((....((((.(.(...((....(((((((.......)))))))..))...)).))))..)))))))((....))..(((....))).............. ( -22.80) >DroYak_CAF1 22900 113 - 1 ACAGCAAAAGCUUUGGAAUGUAAGGGCAAGACAAGAAGUACCAAAUAGGAUGGUACUGAUGAUGGCGUUGCUCUGAAGCUGCAAAAGCAAUGUGUUAGCAAAAGCCCU-AAAAA ...((...(((((..((..((((.(.((...((...(((((((.......)))))))..)).)).).))))))..)))))((....)).........)).........-..... ( -28.50) >consensus ACAGCAAAAGCUUUGGAAUGUAAGGACAAGCCAAGAAGUACCAAAAAGAAUGGUACUGAUGAUGGCGUUGCUCUGAAGCUGCAAAAGCAAUGUGUUAGCAAAAGCCCU_AA_AA ........(((((((((..((((.(.((........(((((((.......))))))).....)).).)))))))))))))((....)).........((....))......... (-23.51 = -23.82 + 0.31)

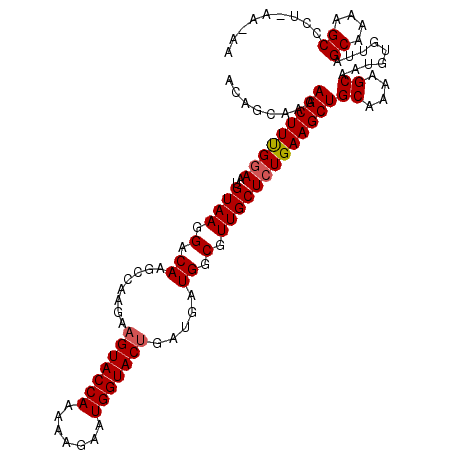
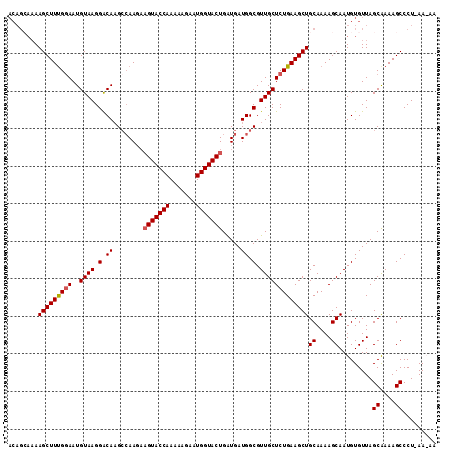
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:36:36 2006