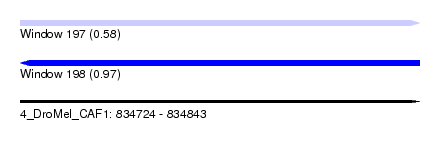
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 834,724 – 834,843 |
| Length | 119 |
| Max. P | 0.971803 |

| Location | 834,724 – 834,843 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -27.38 |
| Consensus MFE | -24.94 |
| Energy contribution | -24.46 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.578680 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 834724 119 + 1281640 AUGCGAAAUUAUUCAAUAAGAA-AGGCUCAAGGCGAAAGAAACUUUUUUCAGAGUCAUCCUUCAGAACUAAGGAGUCAGUCAAUGAAGCGAUGACACAGCGUGUCAUCUGUGUAAUUUGC ..((.(((((((.((....((.-.(((((....(....)...(((..(((.(((......))).)))..))))))))..))........((((((((...)))))))))).))))))))) ( -28.60) >DroSec_CAF1 13818 120 + 1 AUGCGAAAUUAUUCAAUAAGAAAAGGCUCAAGGCGAAAGAAACUUUUUUCAGAGUCAUCCUUCAGAACUAAGGAGUCAGUCAAUGAAGCGAUGACACAGCGUGUCAUCUGUGUAAUUUGC ..((.(((((((.((..........(((((.(((((((((....))))))...(.(.(((((.......)))))).).)))..)).)))((((((((...)))))))))).))))))))) ( -27.80) >DroSim_CAF1 13308 120 + 1 AUGCGAAAUUAUUCAAUAAGAAAAGGCUCAAGGCGAAAGAAACUUUUUUCAGAGUCAUCCUUCAGAACUAAGGAGUCAGUCAAUGAAGCGAUGACACAGCGUGUCAUCUGUGUAAUUUGC ..((.(((((((.((..........(((((.(((((((((....))))))...(.(.(((((.......)))))).).)))..)).)))((((((((...)))))))))).))))))))) ( -27.80) >DroEre_CAF1 13173 120 + 1 AUGCGAAAUUAUUCAAUAAGAAAAGGCUCAACGCAAAAGAAACUUUUUUCAGAGUCAUCCUCCAAUACUAAGGAGUCAGUCAAUGAAGCGAUGACACAGCGUGUCAUCUGUGUAAUUUGC ..((.(((((((.((....((...(((((.....(((((...)))))....)))))...((((........))))))............((((((((...)))))))))).))))))))) ( -26.00) >DroYak_CAF1 16288 120 + 1 AUGCGAAAUUAUUCAAUAAGAAAAGGCUCAACGCAAAAGAAACUUUUUUCAGAGUCAUUCUCCAGAACUAAGGAGUCAGUCAAUGAAGCGAUGACACAGCGUGUCAUCUGUGUAAUUUGC ..((.(((((((.((...(((((((..((.........))..)))))))..(..(((((((((........))))......)))))..)((((((((...)))))))))).))))))))) ( -26.70) >consensus AUGCGAAAUUAUUCAAUAAGAAAAGGCUCAAGGCGAAAGAAACUUUUUUCAGAGUCAUCCUUCAGAACUAAGGAGUCAGUCAAUGAAGCGAUGACACAGCGUGUCAUCUGUGUAAUUUGC ..((.(((((((.((....((...(((((.....(((((...)))))....)))))...((((........))))))............((((((((...)))))))))).))))))))) (-24.94 = -24.46 + -0.48)

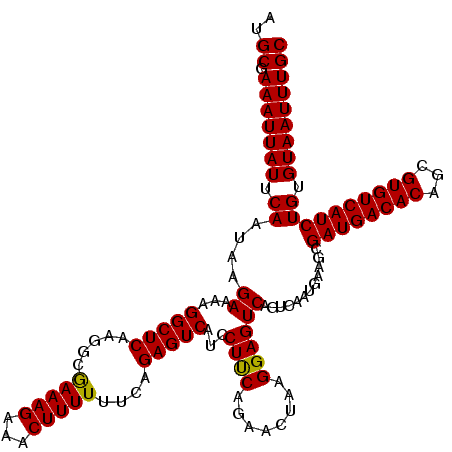
| Location | 834,724 – 834,843 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.17 |
| Mean single sequence MFE | -28.08 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.46 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 834724 119 - 1281640 GCAAAUUACACAGAUGACACGCUGUGUCAUCGCUUCAUUGACUGACUCCUUAGUUCUGAAGGAUGACUCUGAAAAAAGUUUCUUUCGCCUUGAGCCU-UUCUUAUUGAAUAAUUUCGCAU ((((((((..((((((((((...))))))))((((....((((((....))))))..((((((.((((.(....).)))))))))).....))))..-.......))..)))))).)).. ( -28.80) >DroSec_CAF1 13818 120 - 1 GCAAAUUACACAGAUGACACGCUGUGUCAUCGCUUCAUUGACUGACUCCUUAGUUCUGAAGGAUGACUCUGAAAAAAGUUUCUUUCGCCUUGAGCCUUUUCUUAUUGAAUAAUUUCGCAU ((((((((..((((((((((...))))))))((((....((((((....))))))..((((((.((((.(....).)))))))))).....))))..........))..)))))).)).. ( -28.80) >DroSim_CAF1 13308 120 - 1 GCAAAUUACACAGAUGACACGCUGUGUCAUCGCUUCAUUGACUGACUCCUUAGUUCUGAAGGAUGACUCUGAAAAAAGUUUCUUUCGCCUUGAGCCUUUUCUUAUUGAAUAAUUUCGCAU ((((((((..((((((((((...))))))))((((....((((((....))))))..((((((.((((.(....).)))))))))).....))))..........))..)))))).)).. ( -28.80) >DroEre_CAF1 13173 120 - 1 GCAAAUUACACAGAUGACACGCUGUGUCAUCGCUUCAUUGACUGACUCCUUAGUAUUGGAGGAUGACUCUGAAAAAAGUUUCUUUUGCGUUGAGCCUUUUCUUAUUGAAUAAUUUCGCAU ((((((((..((((((((((...))))))))((((..........((((........))))((((.....((((....)))).....))))))))..........))..)))))).)).. ( -27.30) >DroYak_CAF1 16288 120 - 1 GCAAAUUACACAGAUGACACGCUGUGUCAUCGCUUCAUUGACUGACUCCUUAGUUCUGGAGAAUGACUCUGAAAAAAGUUUCUUUUGCGUUGAGCCUUUUCUUAUUGAAUAAUUUCGCAU ((((((((..((((((((((...))))))))((((..........((((........))))((((.....((((....)))).....))))))))..........))..)))))).)).. ( -26.70) >consensus GCAAAUUACACAGAUGACACGCUGUGUCAUCGCUUCAUUGACUGACUCCUUAGUUCUGAAGGAUGACUCUGAAAAAAGUUUCUUUCGCCUUGAGCCUUUUCUUAUUGAAUAAUUUCGCAU ((((((((..((((((((((...))))))))((((....((((((....))))))..((((((.((((.(....).)))))))))).....))))..........))..)))))).)).. (-26.54 = -26.46 + -0.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:49 2006