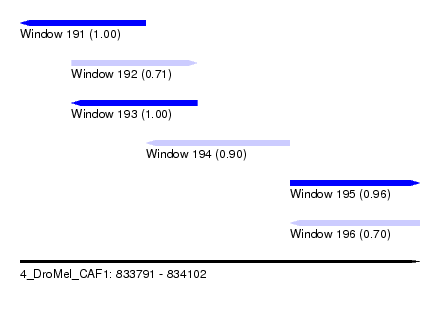
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 833,791 – 834,102 |
| Length | 311 |
| Max. P | 0.999599 |

| Location | 833,791 – 833,889 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -25.56 |
| Energy contribution | -24.84 |
| Covariance contribution | -0.72 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.89 |
| SVM decision value | 3.40 |
| SVM RNA-class probability | 0.999153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 833791 98 - 1281640 GCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCAUGGGGAUGACUAUGAGUAGAAGUGGCAAAGGCGGUGUGGCUGCUGCAAA ...(((((((........(((((((((((((.....)))))))))))))....)))))))....((((.(((.(((.......))).))).))))... ( -28.70) >DroSec_CAF1 12328 98 - 1 GCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCAUGGGGACGACUAUGAGUAGAAGUGGCAAAGGCGCUGUGGCUGCUGCAAA ((.((((...........(((((((((((((.....))))))))))))).(....)))))...(((((..(..((......))..)..)))))))... ( -31.80) >DroSim_CAF1 11813 98 - 1 GCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCAUGGGGACGACUAUGAGUAGAAGUGGCAAAGGCGCUGUGGCUGCUGCAAA ((.((((...........(((((((((((((.....))))))))))))).(....)))))...(((((..(..((......))..)..)))))))... ( -31.80) >DroEre_CAF1 11631 98 - 1 GCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCCUGGGGACGACUAUGAGUAGAAGUGGCGAAGGCGGUGUGGCUGCUGCAAA ((.((((............((((((((((((.....))))))))))))..(....)))))...(((((..((.((....)).))....)))))))... ( -27.40) >DroYak_CAF1 14461 98 - 1 GCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUUCUGGGGACGACUAUGAGUAGAAGUGGCGAAGGCAGUGUGGUUGCUGCAAA ....((((((...((((((((((((((((((.....))))))))))))....))))))(((....))).))))))....(((((......)))))... ( -23.90) >consensus GCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCAUGGGGACGACUAUGAGUAGAAGUGGCAAAGGCGGUGUGGCUGCUGCAAA ...((((............((((((((((((.....))))))))))))..(....)))))....((((.(((.(((.......))).))).))))... (-25.56 = -24.84 + -0.72)

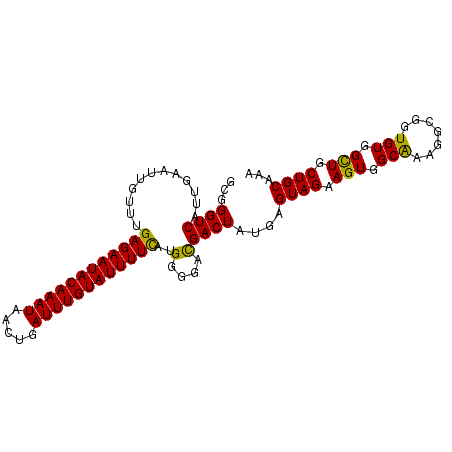
| Location | 833,831 – 833,929 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -16.38 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.712557 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 833831 98 + 1281640 UCAUCCCCAUGAAAAUACAAAUCAGUUAUUUGUAUUCUCAAACAAUUCAAUGACCCGCACGUGCAGUUGCAUCACCCUGCUUACAGUACGUUCCCUUU .........(((.(((((((((.....))))))))).)))................(.((((((.((.(((......)))..)).)))))).)..... ( -18.10) >DroSec_CAF1 12368 98 + 1 UCGUCCCCAUGAAAAUACAAAUCAGUUAUUUGUAUUCUCAAACAAUUCAAUGACCCGCACGUGCAGUUGCAUCACCCUGCUUACAGUACGUUCCCUUU .........(((.(((((((((.....))))))))).)))................(.((((((.((.(((......)))..)).)))))).)..... ( -18.10) >DroSim_CAF1 11853 98 + 1 UCGUCCCCAUGAAAAUACAAAUCAGUUAUUUGUAUUCUCAAACAAUUCAAUGACCCGCACGUGCAGUUGCAUCACCCUGCUUACAGUACGUUCCCUUU .........(((.(((((((((.....))))))))).)))................(.((((((.((.(((......)))..)).)))))).)..... ( -18.10) >DroEre_CAF1 11671 98 + 1 UCGUCCCCAGGAAAAUACAAAUCAGUUAUUUGUAUUCUCAAACAAUUCAAUGACCCGCACGUGCAGUUGCAUCACCCUGCUUACAGUACGCUCCCUUU ..........((.(((((((((.....))))))))).))....................(((((.((.(((......)))..)).)))))........ ( -15.80) >DroYak_CAF1 14501 98 + 1 UCGUCCCCAGAAAAAUACAAAUCAGUUAUUUGUAUUCUCAAACAAUUCAAUGACCCGCACGUGCAGUUGCAUCACCCUGCUCACAGAACGCUCCCUUU .........((..(((((((((.....))))))))).)).................((..((((....))))....(((....)))...))....... ( -11.80) >consensus UCGUCCCCAUGAAAAUACAAAUCAGUUAUUUGUAUUCUCAAACAAUUCAAUGACCCGCACGUGCAGUUGCAUCACCCUGCUUACAGUACGUUCCCUUU ..........((.(((((((((.....))))))))).))....................(((((.((.(((......)))..)).)))))........ (-13.80 = -14.20 + 0.40)

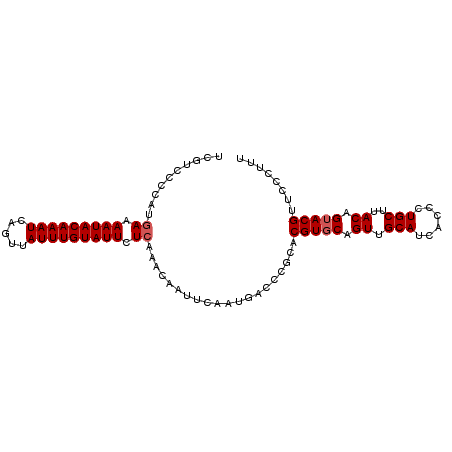
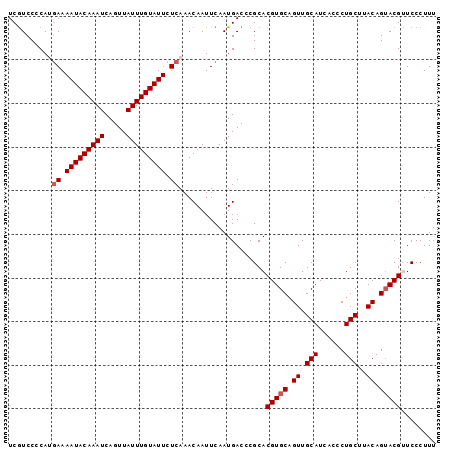
| Location | 833,831 – 833,929 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -27.74 |
| Consensus MFE | -26.90 |
| Energy contribution | -26.42 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.77 |
| SVM RNA-class probability | 0.999599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 833831 98 - 1281640 AAAGGGAACGUACUGUAAGCAGGGUGAUGCAACUGCACGUGCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCAUGGGGAUGA ........(((.((((((((((..(((((...((((....))))..)))))..))))))((((((((((((.....))))))))))))))))..))). ( -26.70) >DroSec_CAF1 12368 98 - 1 AAAGGGAACGUACUGUAAGCAGGGUGAUGCAACUGCACGUGCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCAUGGGGACGA ........(((.((((((((((..(((((...((((....))))..)))))..))))))((((((((((((.....))))))))))))))))..))). ( -28.50) >DroSim_CAF1 11853 98 - 1 AAAGGGAACGUACUGUAAGCAGGGUGAUGCAACUGCACGUGCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCAUGGGGACGA ........(((.((((((((((..(((((...((((....))))..)))))..))))))((((((((((((.....))))))))))))))))..))). ( -28.50) >DroEre_CAF1 11671 98 - 1 AAAGGGAGCGUACUGUAAGCAGGGUGAUGCAACUGCACGUGCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCCUGGGGACGA ...((((........(((((((..(((((...((((....))))..)))))..))))))).((((((((((.....))))))))))))))(....).. ( -26.40) >DroYak_CAF1 14501 98 - 1 AAAGGGAGCGUUCUGUGAGCAGGGUGAUGCAACUGCACGUGCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUUCUGGGGACGA ........((((((..((((((..(((((...((((....))))..)))))..))))))((((((((((((.....))))))))))))...)))))). ( -28.60) >consensus AAAGGGAACGUACUGUAAGCAGGGUGAUGCAACUGCACGUGCGGGUCAUUGAAUUGUUUGAGAAUACAAAUAACUGAUUUGUAUUUUCAUGGGGACGA ........(((.((..((((((..(((((...((((....))))..)))))..))))))((((((((((((.....))))))))))))...)).))). (-26.90 = -26.42 + -0.48)

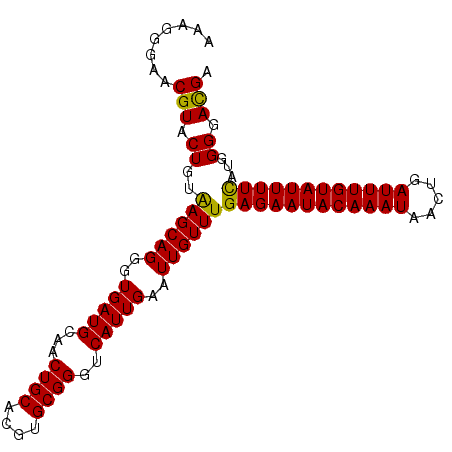
| Location | 833,889 – 834,001 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.83 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -24.03 |
| Energy contribution | -23.91 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 833889 112 - 1281640 UAGCAUUU--------AAAAAAAGGCAAGCGAAUAUAACUUGAACGCUUGUCUUGAUGGACCCUUAAAAUACCAUAUAGAAAAGGGAACGUACUGUAAGCAGGGUGAUGCAACUGCACGU ..(((((.--------.....((((((((((..((.....))..)))))))))).(((..(((((................)))))..))).(((....)))...))))).......... ( -26.99) >DroSec_CAF1 12426 112 - 1 UAGACUUU--------AAAAAAAGGCAAGCGAAUAUAACUUCAACGCUUGUCUUGAUGGACCCUUAAAACAACAUAUAGAAAAGGGAACGUACUGUAAGCAGGGUGAUGCAACUGCACGU (((.....--------.....((((((((((.............)))))))))).(((..(((((................)))))..))).)))...((((..........)))).... ( -25.31) >DroSim_CAF1 11911 112 - 1 UAGACUUU--------AAAAAAAGGCAAGCGAAUAUAACUUCAACGCUUGUCUUGAUGGACCCUUAAAACAACAUAUAGAAAAGGGAACGUACUGUAAGCAGGGUGAUGCAACUGCACGU (((.....--------.....((((((((((.............)))))))))).(((..(((((................)))))..))).)))...((((..........)))).... ( -25.31) >DroEre_CAF1 11729 120 - 1 UAGACUUUUUUUUAAAAAAAAAGGGCAAGCGAAUAUAACUUGAACGCUUGCCUUGAUGGACCCUUAAAAUUCCACAUAGAAAAGGGAGCGUACUGUAAGCAGGGUGAUGCAACUGCACGU (((.((((((((((.......((((((((((..((.....))..))))))))))..((((..........))))..)))))))))).((((.(((....)))....))))..)))..... ( -30.90) >DroYak_CAF1 14559 117 - 1 UAGACUUUA---AAUAAAAAAAAGGCAAGCGAAUAUAACUUGAACGCUUCUCUUGAUGGACCCUUAAAAUACCACAUAGAAAAGGGAGCGUUCUGUGAGCAGGGUGAUGCAACUGCACGU .........---................(((......((..(((((((((.(((.(((................)))....)))))))))))).))..((((..........)))).))) ( -23.39) >consensus UAGACUUU________AAAAAAAGGCAAGCGAAUAUAACUUGAACGCUUGUCUUGAUGGACCCUUAAAAUACCAUAUAGAAAAGGGAACGUACUGUAAGCAGGGUGAUGCAACUGCACGU .....................((((((((((.............)))))))))).(((..(((((................)))))..))).......((((..........)))).... (-24.03 = -23.91 + -0.12)

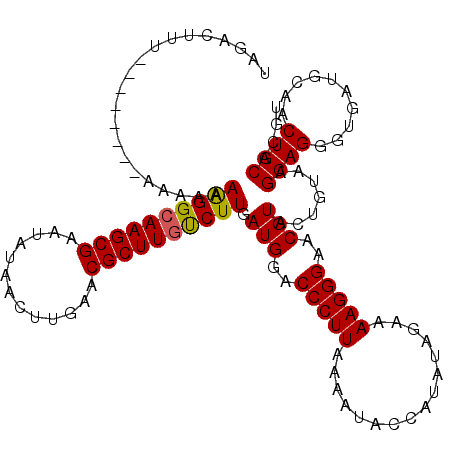
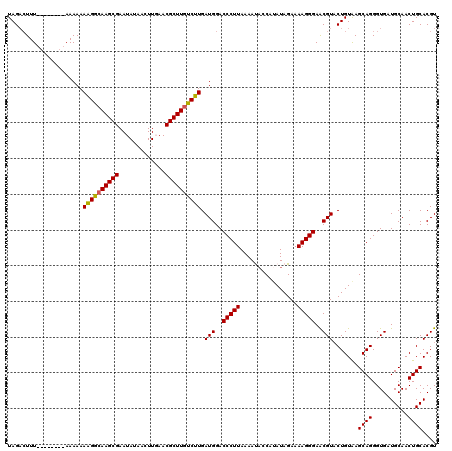
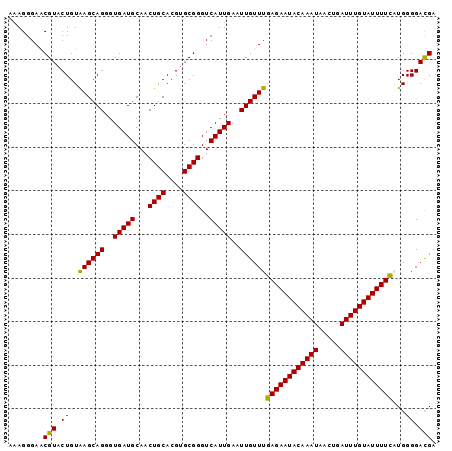
| Location | 834,001 – 834,102 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.40 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -19.48 |
| Energy contribution | -19.32 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.63 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 834001 101 + 1281640 UCGACA-AAAACGUUGAAGAAUCUCAGGUGUGUUCCCACAGAGGAGAGGAUAUGCUA---CCUCUGGUGAGAUAAGCCC--GGGGGGUUCACCCAAAAAAAAGAA-------U------G (((((.-.....))))).(((((((.((.((..((.((((((((((........)).---))))).))).))...))))--..)))))))...............-------.------. ( -29.40) >DroSec_CAF1 12538 103 + 1 UCGACA-AAAACGUUGAAAAAUCUCAGGUGUGUUCCCACAGAGGAGAGGACACGCUACCACCUCUGGUGGGAUAAGCCC--AGGGGGUUCACCCAAA-CAAAGAA-------U------G (((((.-.....))))).........((((.(..(((.((((((...((........)).)))))).((((.....)))--))))..).))))....-.......-------.------. ( -33.00) >DroSim_CAF1 12023 103 + 1 UCGACA-AAAACGUUGAAAAAUCUCAGGUGUGUUCCCACAGAGGAGAGGACACGCUACCACCUCUGGUGGGAUAAGCCC--AGGGGGUUCACCCAAA-CAAAGAA-------U------G (((((.-.....))))).........((((.(..(((.((((((...((........)).)))))).((((.....)))--))))..).))))....-.......-------.------. ( -33.00) >DroEre_CAF1 11849 111 + 1 UCAGCCAAAAAUGUGGAAGAAUCACAGGUGUGUUUCCACAUAGGACUUG----GCU---AGCUCUGGUGGGAUAGACCCGGAGGGGGUUCACCAAAA-CAAAGAAAA-AGUAUUUCAGCG ..((((((..(((((((((...(((....)))))))))))).....)))----)))---.(((.(((((((.....(((....))).)))))))...-....((((.-....))))))). ( -34.40) >DroYak_CAF1 14676 110 + 1 UCGGCG-AAAGUGUGGAAGAUUCACAGGUGUGUUCCCAUAUAAGAUAUG----GCU---AGCUCUGGUGGGAUAAGCCCUAAGGGAGUUCAUUCAAA-CAAAGAAAAAAGAAUUUCA-CA ...((.-...))(((((((((((..(((.((..((((((...(((....----...---...))).))))))...)))))....)))))..(((...-....))).......)))))-). ( -25.30) >consensus UCGACA_AAAACGUUGAAGAAUCUCAGGUGUGUUCCCACAGAGGAGAGGA_A_GCUA__ACCUCUGGUGGGAUAAGCCC__AGGGGGUUCACCCAAA_CAAAGAA_______U______G (((((.......))))).........((((...((((((...((((...............)))).))))))..(((((.....)))))))))........................... (-19.48 = -19.32 + -0.16)

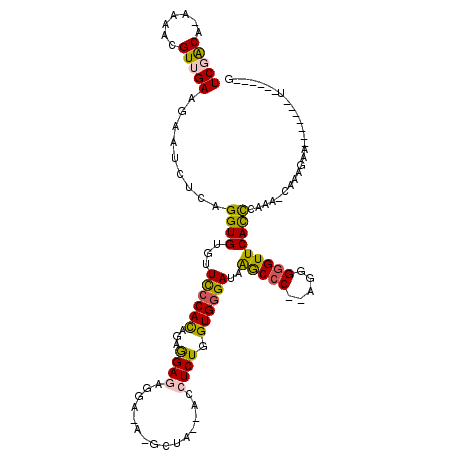
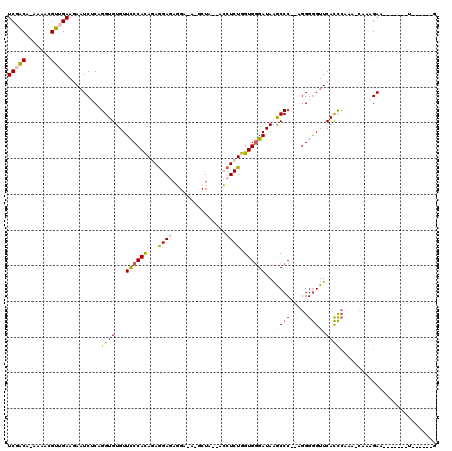
| Location | 834,001 – 834,102 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.40 |
| Mean single sequence MFE | -28.64 |
| Consensus MFE | -16.21 |
| Energy contribution | -17.93 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 834001 101 - 1281640 C------A-------UUCUUUUUUUUGGGUGAACCCCCC--GGGCUUAUCUCACCAGAGG---UAGCAUAUCCUCUCCUCUGUGGGAACACACCUGAGAUUCUUCAACGUUUU-UGUCGA .------.-------.......(((..((((..(((...--)))....((((((.(((((---.((........)))))))))))))...))))..)))....((.(((....-))).)) ( -26.90) >DroSec_CAF1 12538 103 - 1 C------A-------UUCUUUG-UUUGGGUGAACCCCCU--GGGCUUAUCCCACCAGAGGUGGUAGCGUGUCCUCUCCUCUGUGGGAACACACCUGAGAUUUUUCAACGUUUU-UGUCGA .------.-------.......-((..((((..(((...--)))....((((((.(((((.((........))...)))))))))))...))))..)).....((.(((....-))).)) ( -31.10) >DroSim_CAF1 12023 103 - 1 C------A-------UUCUUUG-UUUGGGUGAACCCCCU--GGGCUUAUCCCACCAGAGGUGGUAGCGUGUCCUCUCCUCUGUGGGAACACACCUGAGAUUUUUCAACGUUUU-UGUCGA .------.-------.......-((..((((..(((...--)))....((((((.(((((.((........))...)))))))))))...))))..)).....((.(((....-))).)) ( -31.10) >DroEre_CAF1 11849 111 - 1 CGCUGAAAUACU-UUUUCUUUG-UUUUGGUGAACCCCCUCCGGGUCUAUCCCACCAGAGCU---AGC----CAAGUCCUAUGUGGAAACACACCUGUGAUUCUUCCACAUUUUUGGCUGA ....((((....-.))))...(-((((((((.((((.....))))......)))))))))(---(((----((((....((((((((.(((....)))....)))))))).)))))))). ( -36.60) >DroYak_CAF1 14676 110 - 1 UG-UGAAAUUCUUUUUUCUUUG-UUUGAAUGAACUCCCUUAGGGCUUAUCCCACCAGAGCU---AGC----CAUAUCUUAUAUGGGAACACACCUGUGAAUCUUCCACACUUU-CGCCGA .(-(((((.............(-(((....))))((((.((.((((..((......))...---)))----)........)).)))).(((....)))............)))-)))... ( -17.50) >consensus C______A_______UUCUUUG_UUUGGGUGAACCCCCU__GGGCUUAUCCCACCAGAGGU__UAGC_U_UCCUCUCCUCUGUGGGAACACACCUGAGAUUCUUCAACGUUUU_UGUCGA .......................((((((((..(((.....)))....((((((.(((((................)))))))))))...))))))))...................... (-16.21 = -17.93 + 1.72)

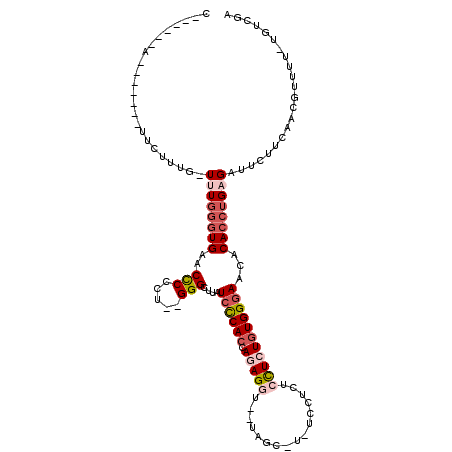
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:46 2006