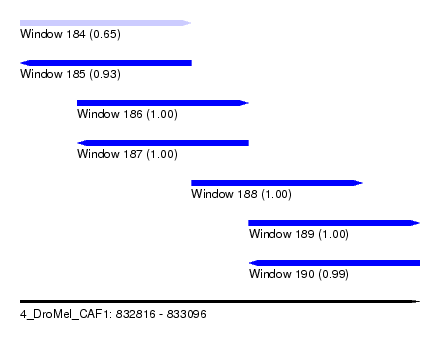
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 832,816 – 833,096 |
| Length | 280 |
| Max. P | 0.999997 |

| Location | 832,816 – 832,936 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -27.12 |
| Energy contribution | -27.08 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 832816 120 + 1281640 CUCUUAUUGCCCAUAUCCUUGGUAAAAGUCGCCGUCCUGGAUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGAGUGAAUACACCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGG ..........(((.((((.((((.......))))....))))..)))......((((((...((((.....((((((.(((....))).))))))))))...)))))).(((.....))) ( -31.40) >DroSec_CAF1 11353 120 + 1 CUUUUAUUGCCCAUAUCCUUGGUAGAAGUCACCGUCCUGGAUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGAGUGAAUACGCCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGG ((((((....(((......)))))))))........(..(((..(((......((((((...((((.....((((((.(((....))).))))))))))...)))))).)))..)))..) ( -31.60) >DroSim_CAF1 10839 120 + 1 CUUUUAUUGCCCAUAUCCUUGGUAGAAGUCACCGUCCUGGAUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGAGUGAAUACGCCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGG ((((((....(((......)))))))))........(..(((..(((......((((((...((((.....((((((.(((....))).))))))))))...)))))).)))..)))..) ( -31.60) >DroEre_CAF1 10669 119 + 1 CUUCUAUUGCCCAUAUCCUUGGUAAAAAUCACCGUCCG-GGUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGUGUGAAUACACCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGG ..((((..((((.......((((.......))))...)-)))..)))).....((((((...((((.....((((((.(((....))).))))))))))...)))))).(((.....))) ( -32.20) >DroYak_CAF1 13491 120 + 1 CCUUUAUUGCCCAUAUCCUUGGUAAAAGUCACAGUCCUGAAUUUUGGAUCUACGUCACAUCCGAAGGCUACAACUUGUGUGAAUACACCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGG ..........(((.((...(((...........((((........))))....((((((...(((((((......((.(((....))).))))).))))...)))))).)))..)).))) ( -24.00) >consensus CUUUUAUUGCCCAUAUCCUUGGUAAAAGUCACCGUCCUGGAUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGAGUGAAUACACCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGG ..........(((.((...(((...........((((........))))....((((((...((((.....((((((.(((....))).))))))))))...)))))).)))..)).))) (-27.12 = -27.08 + -0.04)

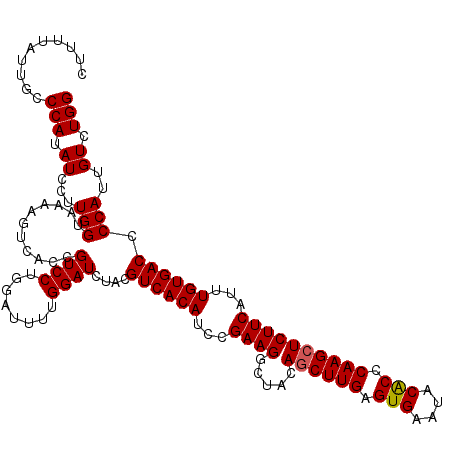
| Location | 832,816 – 832,936 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -34.66 |
| Consensus MFE | -31.92 |
| Energy contribution | -32.56 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 832816 120 - 1281640 CCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGUGUAUUCACUCAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAUCCAGGACGGCGACUUUUACCAAGGAUAUGGGCAAUAAGAG (((..(..(((.((((((..(((((((((((((((....)))))))))).....)))))..))))))((...(((........)))..))........)))..)...))).......... ( -37.80) >DroSec_CAF1 11353 120 - 1 CCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGCGUAUUCACUCAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAUCCAGGACGGUGACUUCUACCAAGGAUAUGGGCAAUAAAAG (((..(..(((.((((((..(((((((((((((.(....).)))))))).....)))))..)))))).(((((((........))).(....).)))))))..)...))).......... ( -35.30) >DroSim_CAF1 10839 120 - 1 CCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGCGUAUUCACUCAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAUCCAGGACGGUGACUUCUACCAAGGAUAUGGGCAAUAAAAG (((..(..(((.((((((..(((((((((((((.(....).)))))))).....)))))..)))))).(((((((........))).(....).)))))))..)...))).......... ( -35.30) >DroEre_CAF1 10669 119 - 1 CCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGUGUAUUCACACAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAACC-CGGACGGUGAUUUUUACCAAGGAUAUGGGCAAUAGAAG (((.....((((((((((..(((((((((((.(((....))).)))))).....)))))..))))))(......)....))-))...(((((...))))).......))).......... ( -33.60) >DroYak_CAF1 13491 120 - 1 CCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGUGUAUUCACACAAGUUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAUUCAGGACUGUGACUUUUACCAAGGAUAUGGGCAAUAAAGG (((..(..(((.((((((..(((((((((((.(((....))).)))))).....)))))..)))))).(((.(((........)))))).........)))..)...))).......... ( -31.30) >consensus CCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGUGUAUUCACUCAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAUCCAGGACGGUGACUUUUACCAAGGAUAUGGGCAAUAAAAG (((..(..(((.((((((..(((((((((((((((....)))))))))).....)))))..)))))).....(((........)))............)))..)...))).......... (-31.92 = -32.56 + 0.64)

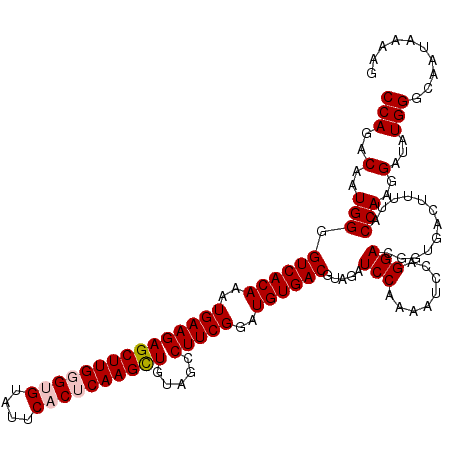
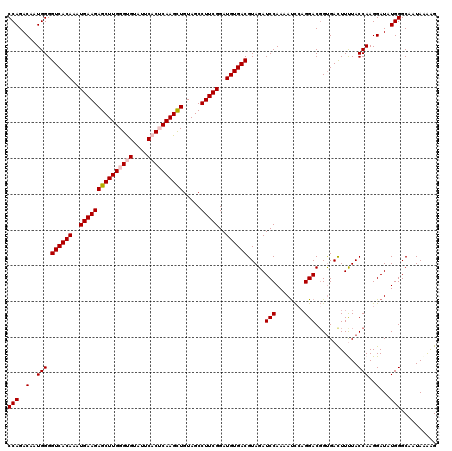
| Location | 832,856 – 832,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -34.60 |
| Energy contribution | -34.56 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.96 |
| SVM RNA-class probability | 0.999729 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 832856 120 + 1281640 AUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGAGUGAAUACACCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGGCAGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGAAUGU ...((((......((((((...((((.....((((((.(((....))).))))))))))...))))))((((((((((....)).))))))))...........))))............ ( -36.10) >DroSec_CAF1 11393 120 + 1 AUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGAGUGAAUACGCCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGGCGGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGAAUGU ...((((......((((((...((((.....((((((.(((....))).))))))))))...))))))((((((((((....)).))))))))...........))))............ ( -35.20) >DroSim_CAF1 10879 120 + 1 AUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGAGUGAAUACGCCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGGCGGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGAAUGU ...((((......((((((...((((.....((((((.(((....))).))))))))))...))))))((((((((((....)).))))))))...........))))............ ( -35.20) >DroEre_CAF1 10708 120 + 1 GUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGUGUGAAUACACCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGGCAGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGACUGU ((.((((......((((((...((((.....((((((.(((....))).))))))))))...))))))((((((((((....)).))))))))...........)))).))......... ( -38.00) >DroYak_CAF1 13531 120 + 1 AUUUUGGAUCUACGUCACAUCCGAAGGCUACAACUUGUGUGAAUACACCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGGCAGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGACUGU ...((((......((((((...(((((((......((.(((....))).))))).))))...))))))((((((((((....)).))))))))...........))))............ ( -31.50) >consensus AUUUUGGAUCUACGUCACAUCCGAAGGCUACAGCUUGAGUGAAUACACCCAAGCUCUUCAUUUGUGACCCCAUUGUCUGGCAGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGAAUGU ...((((......((((((...((((.....((((((.(((....))).))))))))))...))))))((((((((((....)).))))))))...........))))............ (-34.60 = -34.56 + -0.04)

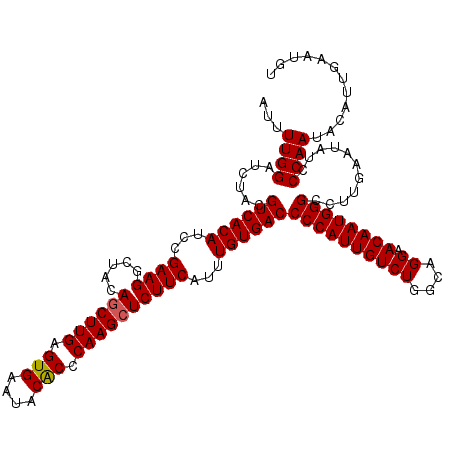
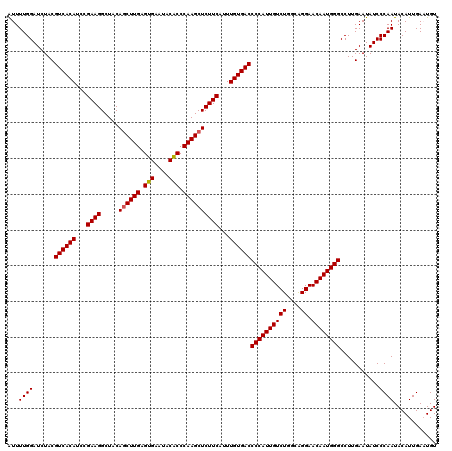
| Location | 832,856 – 832,976 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.33 |
| Mean single sequence MFE | -41.56 |
| Consensus MFE | -40.72 |
| Energy contribution | -41.36 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.04 |
| SVM RNA-class probability | 0.999970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 832856 120 - 1281640 ACAUUCAAUGUAUUGGGAUAUUCAAGGCCCAUUGUUCCUGCCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGUGUAUUCACUCAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAU ...............((((...(....(((((((((.......)))))))))((((((..(((((((((((((((....)))))))))).....)))))..)))))))...))))..... ( -45.00) >DroSec_CAF1 11393 120 - 1 ACAUUCAAUGUAUUGGGAUAUUCAAGGCCCAUUGUUCCCGCCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGCGUAUUCACUCAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAU ...............((((...(....(((((((((.......)))))))))((((((..(((((((((((((.(....).)))))))).....)))))..)))))))...))))..... ( -41.10) >DroSim_CAF1 10879 120 - 1 ACAUUCAAUGUAUUGGGAUAUUCAAGGCCCAUUGUUCCCGCCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGCGUAUUCACUCAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAU ...............((((...(....(((((((((.......)))))))))((((((..(((((((((((((.(....).)))))))).....)))))..)))))))...))))..... ( -41.10) >DroEre_CAF1 10708 120 - 1 ACAGUCAAUGUAUUGGGAUAUUCAAGGCCCAUUGUUCCUGCCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGUGUAUUCACACAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAC .((((......))))((((...(....(((((((((.......)))))))))((((((..(((((((((((.(((....))).)))))).....)))))..)))))))...))))..... ( -41.50) >DroYak_CAF1 13531 120 - 1 ACAGUCAAUGUAUUGGGAUAUUCAAGGCCCAUUGUUCCUGCCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGUGUAUUCACACAAGUUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAU .((((......))))((((...(....(((((((((.......)))))))))((((((..(((((((((((.(((....))).)))))).....)))))..)))))))...))))..... ( -39.10) >consensus ACAUUCAAUGUAUUGGGAUAUUCAAGGCCCAUUGUUCCUGCCAGACAAUGGGGUCACAAAUGAAGAGCUUGGGUGUAUUCACUCAAGCUGUAGCCUUCGGAUGUGACGUAGAUCCAAAAU ...............((((...(....(((((((((.......)))))))))((((((..(((((((((((((((....)))))))))).....)))))..)))))))...))))..... (-40.72 = -41.36 + 0.64)

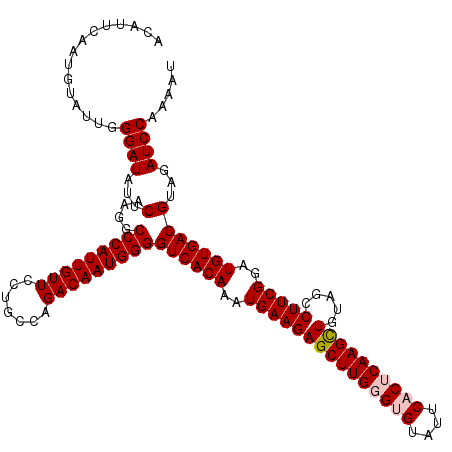
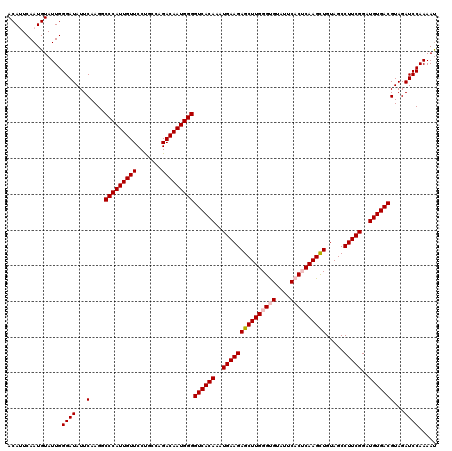
| Location | 832,936 – 833,056 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.83 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -28.92 |
| Energy contribution | -29.08 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 832936 120 + 1281640 CAGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGAAUGUGGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUUCUAUUUUGUCUUUUGUUGUCAGAGAUUUAAU ..........(.((((((.......))).((((...))))))).)..((((((((((((((....(((((..((..((......))...))...)))))))))))))))))))....... ( -26.00) >DroSec_CAF1 11473 120 + 1 CGGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGAAUGUGGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUCAGAGAUUUAAU .((((.(((......)))....))))..(((((...)))))......((((((((((((((....(((((..((((((......).)))))...)))))))))))))))))))....... ( -32.90) >DroSim_CAF1 10959 120 + 1 CGGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGAAUGUGGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUCAGAGAUUUAAU .((((.(((......)))....))))..(((((...)))))......((((((((((((((....(((((..((((((......).)))))...)))))))))))))))))))....... ( -32.90) >DroEre_CAF1 10788 120 + 1 CAGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGACUGUGGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUUAGAGAUUUAAU (((...(((((....(((.......)))..))))).)))........((((((((((((((....(((((..((((((......).)))))...)))))))))))))))))))....... ( -30.50) >DroYak_CAF1 13611 120 + 1 CAGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGACUGUGGCGCUACUUUGGCAACAAAACAUUAACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUUAGAGAUUUAAU (((...(((((....(((.......)))..))))).)))........((((((((((((((.....((((..((((((......).)))))...)))).))))))))))))))....... ( -26.80) >consensus CAGGAACAAUGGGCCUUGAAUAUCCCAAUACAUUGAAUGUGGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUCAGAGAUUUAAU ..........(.((((((.......))).(((.....)))))).)..((((((((((((((....(((((..((((((......).)))))...)))))))))))))))))))....... (-28.92 = -29.08 + 0.16)

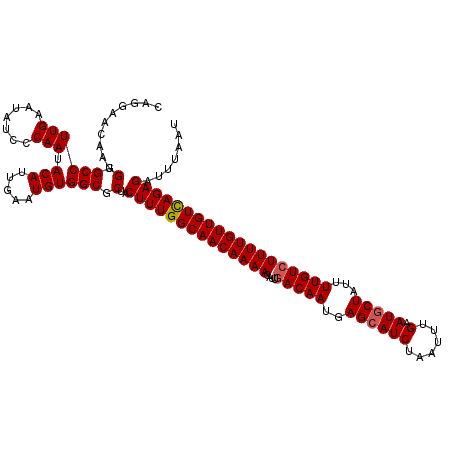
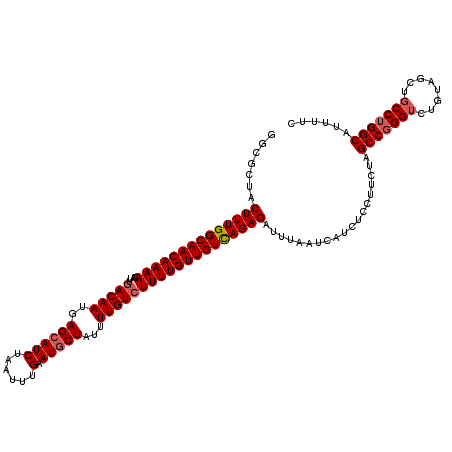
| Location | 832,976 – 833,096 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -34.63 |
| Energy contribution | -35.24 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.15 |
| Structure conservation index | 1.01 |
| SVM decision value | 6.15 |
| SVM RNA-class probability | 0.999997 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 832976 120 + 1281640 GGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUUCUAUUUUGUCUUUUGUUGUCAGAGAUUUAAUCAUAUCCUUCUGGCCGGGUCUGUAGCUGCCUGGCAUUUUC .......((((((((((((((....(((((..((..((......))...))...)))))))))))))))))))...................(((((((........)))))))...... ( -33.50) >DroSec_CAF1 11513 120 + 1 GGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUCAGAGAUUUAAUCAUCUCCUUCUAGCCGGGUCUGUAGCUGCCUGGCAUUUUC .......((((((((((((((....(((((..((((((......).)))))...)))))))))))))))))))...................(((((((........)))))))...... ( -39.50) >DroSim_CAF1 10999 120 + 1 GGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUCAGAGAUUUAAUCAUCUCCUUCUAGCCGGGUCUGUAGCUGCCUGGCAUUUUC .......((((((((((((((....(((((..((((((......).)))))...)))))))))))))))))))...................(((((((........)))))))...... ( -39.50) >DroEre_CAF1 10828 110 + 1 GGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUUAGAGAUUUAAUCAUCUCCUUCUAGCCGGG----------CCUGGCAUUUUC (((((((...(((((((((((....(((((..((((((......).)))))...))))))))))))))))(((((......)))))....))))...)----------)).......... ( -30.70) >DroYak_CAF1 13651 120 + 1 GGCGCUACUUUGGCAACAAAACAUUAACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUUAGAGAUUUAAUCAUCUCCUUCUAGCCAGGUUUGCAGCUUCCCGGCAGUUUC ...(((.((((((((((((((.....((((..((((((......).)))))...)))).))))))))))))))...................(((.((..........)).))))))... ( -28.30) >consensus GGCGCUACUUUGGCAACAAAACAUUGACAAUGAGCAUCUAAUUUGAAUGCUAUUUUGUCUUUUGUUGUCAGAGAUUUAAUCAUCUCCUUCUAGCCGGGUCUGUAGCUGCCUGGCAUUUUC .......((((((((((((((....(((((..((((((......).)))))...)))))))))))))))))))...................(((((((........)))))))...... (-34.63 = -35.24 + 0.61)

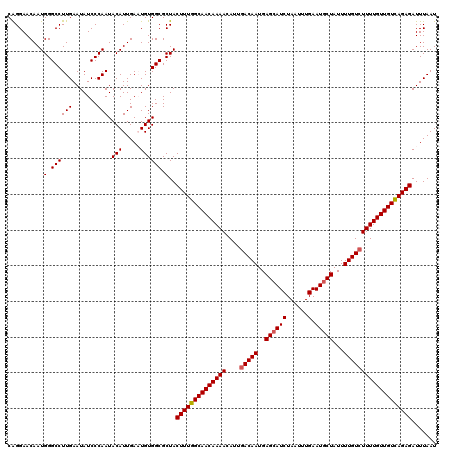
| Location | 832,976 – 833,096 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.08 |
| Mean single sequence MFE | -30.14 |
| Consensus MFE | -24.92 |
| Energy contribution | -26.44 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993570 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 832976 120 - 1281640 GAAAAUGCCAGGCAGCUACAGACCCGGCCAGAAGGAUAUGAUUAAAUCUCUGACAACAAAAGACAAAAUAGAAUUCAAAUUAGAUGCUCAUUGUCAAUGUUUUGUUGCCAAAGUAGCGCC ..........(((.(((((.........((((..(((........))))))).(((((((((((((...((...((......))..))..)))))....)))))))).....)))))))) ( -25.00) >DroSec_CAF1 11513 120 - 1 GAAAAUGCCAGGCAGCUACAGACCCGGCUAGAAGGAGAUGAUUAAAUCUCUGACAACAAAAGACAAAAUAGCAUUCAAAUUAGAUGCUCAUUGUCAAUGUUUUGUUGCCAAAGUAGCGCC ..........(((.(((((..............((((((......))))))(.(((((((((((((...((((((.......))))))..)))))....)))))))).)...)))))))) ( -33.80) >DroSim_CAF1 10999 120 - 1 GAAAAUGCCAGGCAGCUACAGACCCGGCUAGAAGGAGAUGAUUAAAUCUCUGACAACAAAAGACAAAAUAGCAUUCAAAUUAGAUGCUCAUUGUCAAUGUUUUGUUGCCAAAGUAGCGCC ..........(((.(((((..............((((((......))))))(.(((((((((((((...((((((.......))))))..)))))....)))))))).)...)))))))) ( -33.80) >DroEre_CAF1 10828 110 - 1 GAAAAUGCCAGG----------CCCGGCUAGAAGGAGAUGAUUAAAUCUCUAACAACAAAAGACAAAAUAGCAUUCAAAUUAGAUGCUCAUUGUCAAUGUUUUGUUGCCAAAGUAGCGCC ..........((----------(...((((...((((((......))))))..(((((((((((((...((((((.......))))))..)))))....))))))))......))))))) ( -28.00) >DroYak_CAF1 13651 120 - 1 GAAACUGCCGGGAAGCUGCAAACCUGGCUAGAAGGAGAUGAUUAAAUCUCUAACAACAAAAGACAAAAUAGCAUUCAAAUUAGAUGCUCAUUGUUAAUGUUUUGUUGCCAAAGUAGCGCC ....((((((((..........)))))).))..((((((......))))))..(((((((((((((...((((((.......))))))..)))))....))))))))............. ( -30.10) >consensus GAAAAUGCCAGGCAGCUACAGACCCGGCUAGAAGGAGAUGAUUAAAUCUCUGACAACAAAAGACAAAAUAGCAUUCAAAUUAGAUGCUCAUUGUCAAUGUUUUGUUGCCAAAGUAGCGCC ..........(((.(((((..............((((((......))))))..(((((((((((((...((((((.......))))))..)))))....)))))))).....)))))))) (-24.92 = -26.44 + 1.52)

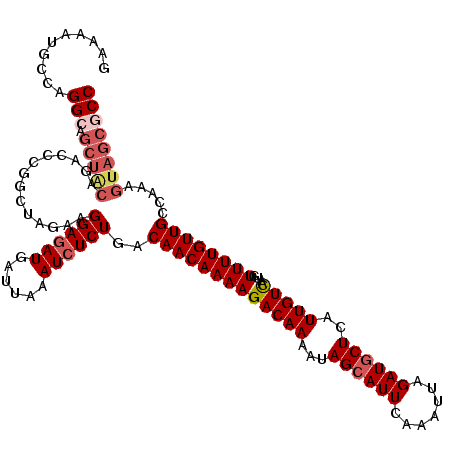
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:23 2006