| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 823,448 – 823,547 |
| Length | 99 |
| Max. P | 0.624690 |

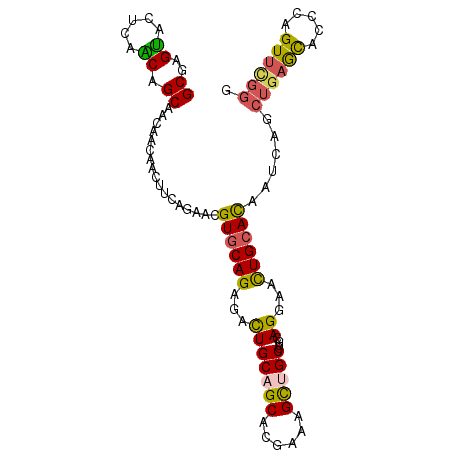
| Location | 823,448 – 823,547 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -18.50 |
| Energy contribution | -17.53 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542070 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 823448 99 + 1281640 CCCGUACUGGCUGCUCAGAUGAUUGUGCAAUUCCUGAAGGCAGCUUUCGUGCUGCAGUCUCUGCACGUUCUGAAGUUGUUGUUGCUGUUGAGCACUCGC ........(..(((((((..((.((((((......((..(((((......)))))..))..)))))).))...(((.......))).)))))))..).. ( -29.20) >DroPse_CAF1 2875 99 + 1 CCCGAACUGUGUAUUCAGCUGAUGAUGCAGCUCCUGCAGGCUACUCUCUUGCUGCAGCCUCUGCACAUUUUGGAGUUGCUGCUGCUGCUGAAUACUUGC ..........(((((((((.(.....((((((((.(((((.......)))))(((((...)))))......)))))))).....).))))))))).... ( -36.70) >DroEre_CAF1 2232 99 + 1 UCCGAACUGGCUGCUCAGUUGAUUGUGCAAUUCCUGAAGGCAGCUUUCGUGCUGCAGUCUCUGCACGUUCUGAAGGCGUUGUUGCUGUUUAGCACUCGC ...(((..((((((((((..((((....)))).))))..)))))))))((((((((((....(((((((.....)))).))).)))))..))))).... ( -29.50) >DroWil_CAF1 2306 96 + 1 CGAAAACUGAGCACUAAGCUGCUGGUGCAGCUCCUGCAGGCAGCUCUCCUGCUGCAAUCGCUGGACAUUGUGCAGUUG---CUGCUGCUGAAUGCUAGC .((.......(((...((((((....))))))..)))..(((((......)))))..))(((((.((((..(((((..---..)))))..))))))))) ( -39.30) >DroYak_CAF1 1897 99 + 1 CCCAAACUGGCUGCUCAGUUGAUUGUGCAAUUCCUGAAGGCAGCUUUCGUGCUGCAGUCUCUGCACGUUCUGAAGGUGUUGUUGCUGUUGAGCACUCGC ........((((((((((..((((....)))).))))..))))))...((((((((((..(.((((.(.....).)))).)..)))))..))))).... ( -29.40) >DroPer_CAF1 2506 99 + 1 CCCGAACUGUGUAUUCAGCUGAUGAUGCAGCUCCUGCAGGCUACUCUCUUGCUGCAGCCUCUGCACAUUUUGGAGUUGCUGCUGCUGCUGAAUACUUGC ..........(((((((((.(.....((((((((.(((((.......)))))(((((...)))))......)))))))).....).))))))))).... ( -36.70) >consensus CCCGAACUGGCUACUCAGCUGAUGGUGCAACUCCUGAAGGCAGCUCUCGUGCUGCAGUCUCUGCACAUUCUGAAGUUGUUGCUGCUGCUGAACACUCGC ...........((((((((.(.....(((((..((....(((((......)))))....((..........))))..)))))..).))))))))..... (-18.50 = -17.53 + -0.97)

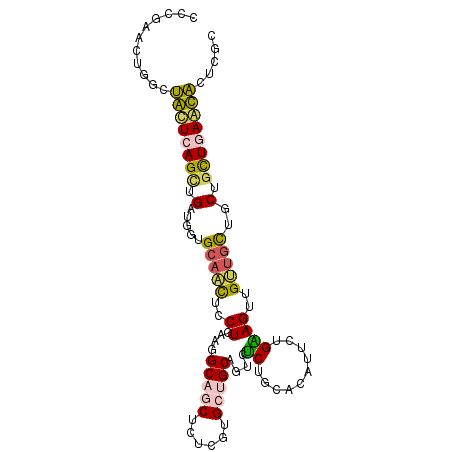
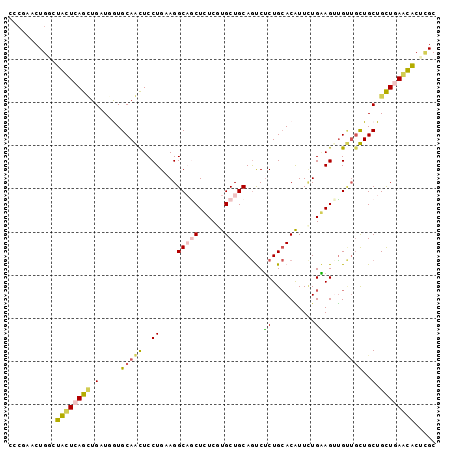
| Location | 823,448 – 823,547 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -17.42 |
| Energy contribution | -16.78 |
| Covariance contribution | -0.63 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 823448 99 - 1281640 GCGAGUGCUCAACAGCAACAACAACUUCAGAACGUGCAGAGACUGCAGCACGAAAGCUGCCUUCAGGAAUUGCACAAUCAUCUGAGCAGCCAGUACGGG ((...((((....)))).........(((((..((((((...(((..((((....).)))...)))...)))))).....)))))))..((.....)). ( -25.00) >DroPse_CAF1 2875 99 - 1 GCAAGUAUUCAGCAGCAGCAGCAACUCCAAAAUGUGCAGAGGCUGCAGCAAGAGAGUAGCCUGCAGGAGCUGCAUCAUCAGCUGAAUACACAGUUCGGG ....(((((((((.((.(((((..((.((.....)).))..))))).))..((((((((((.....).))))).)).)).))))))))).......... ( -35.70) >DroEre_CAF1 2232 99 - 1 GCGAGUGCUAAACAGCAACAACGCCUUCAGAACGUGCAGAGACUGCAGCACGAAAGCUGCCUUCAGGAAUUGCACAAUCAACUGAGCAGCCAGUUCGGA (((..((((....))))....))).........((((((...(((..((((....).)))...)))...))))))......((((((.....)))))). ( -29.00) >DroWil_CAF1 2306 96 - 1 GCUAGCAUUCAGCAGCAG---CAACUGCACAAUGUCCAGCGAUUGCAGCAGGAGAGCUGCCUGCAGGAGCUGCACCAGCAGCUUAGUGCUCAGUUUUCG (((.(((.((.((((...---...)))).....(.....))).))))))..((((((((...(((.(((((((....)))))))..))).)))))))). ( -37.90) >DroYak_CAF1 1897 99 - 1 GCGAGUGCUCAACAGCAACAACACCUUCAGAACGUGCAGAGACUGCAGCACGAAAGCUGCCUUCAGGAAUUGCACAAUCAACUGAGCAGCCAGUUUGGG .....((((....))))......((........((((((...(((..((((....).)))...)))...))))))....(((((......))))).)). ( -23.80) >DroPer_CAF1 2506 99 - 1 GCAAGUAUUCAGCAGCAGCAGCAACUCCAAAAUGUGCAGAGGCUGCAGCAAGAGAGUAGCCUGCAGGAGCUGCAUCAUCAGCUGAAUACACAGUUCGGG ....(((((((((.((.(((((..((.((.....)).))..))))).))..((((((((((.....).))))).)).)).))))))))).......... ( -35.70) >consensus GCGAGUACUCAACAGCAACAACAACUUCAGAACGUGCAGAGACUGCAGCACGAAAGCUGCCUGCAGGAACUGCACAAUCAGCUGAGCACCCAGUUCGGG ((..((.....)).)).................((((((...(((((((......)))))....))...))))))......((((((.....)))))). (-17.42 = -16.78 + -0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:00 2006