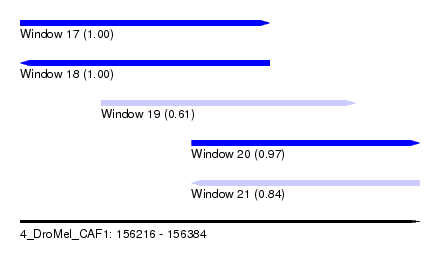
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 156,216 – 156,384 |
| Length | 168 |
| Max. P | 0.999483 |

| Location | 156,216 – 156,321 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 71.35 |
| Mean single sequence MFE | -26.80 |
| Consensus MFE | -13.03 |
| Energy contribution | -13.04 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.49 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 156216 105 + 1281640 AAUAAAUAUGUAUA------UGCAUUUGUAUGUAUAUGAAGCAACAAAUCUGUUU-UCAGCGUUGUUGUAUUAAUAUAU----GCAUACAUACAUGUAUUGAUCAGCACAAGCCAG ......((((((((------(((....)))))))))))..(((((((..(((...-.)))..)))))))((((((((((----(........)))))))))))..((....))... ( -27.10) >DroEre_CAF1 18144 116 + 1 AGUAUAUAUGCAUAUAUGAUUGCAUAUGUAGGUAUAUGAAACAACGAAUCUGUUUGACAGUGGUGCUAUAUUAAUAUGUACACACAUAUGUACACGUAUUGAUCAGCACAUGCCAG ..((((((((((........))))))))))(((((.(.(((((.......))))).).....(((((..((((((((((..(((....)))..)))))))))).)))))))))).. ( -33.40) >DroYak_CAF1 18462 96 + 1 -------GUGCA-------------ACGUAUGCAUAUAAAACAACGAAUCUGUUUGACAGAGUUGCUGUAUUAAUAUGAACAUACAUACAUACAUGUAUUGAUCAGCAAAUGCCAG -------.(((.-------------..((((((((((((.(((.(((.((((.....)))).))).))).)).)))))..)))))(((((....)))))......)))........ ( -19.90) >consensus A_UA_AUAUGCAUA______UGCAUAUGUAUGUAUAUGAAACAACGAAUCUGUUUGACAGAGUUGCUGUAUUAAUAUGUACA_ACAUACAUACAUGUAUUGAUCAGCACAUGCCAG ...............................((((...(((((.......))))).......((((((.((((((((((..............))))))))))))))))))))... (-13.03 = -13.04 + 0.01)

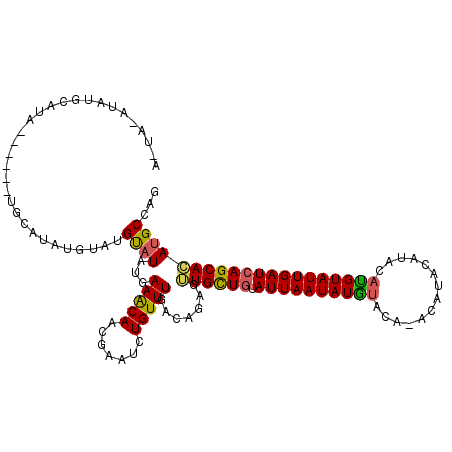
| Location | 156,216 – 156,321 |
|---|---|
| Length | 105 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.35 |
| Mean single sequence MFE | -22.66 |
| Consensus MFE | -14.94 |
| Energy contribution | -15.73 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.64 |
| SVM RNA-class probability | 0.999483 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 156216 105 - 1281640 CUGGCUUGUGCUGAUCAAUACAUGUAUGUAUGC----AUAUAUUAAUACAACAACGCUGA-AAACAGAUUUGUUGCUUCAUAUACAUACAAAUGCA------UAUACAUAUUUAUU ..(((....)))....((((.((((.(((((((----((((((.....((((((..(((.-...)))..))))))....)))))........))))------)))).)))).)))) ( -19.90) >DroEre_CAF1 18144 116 - 1 CUGGCAUGUGCUGAUCAAUACGUGUACAUAUGUGUGUACAUAUUAAUAUAGCACCACUGUCAAACAGAUUCGUUGUUUCAUAUACCUACAUAUGCAAUCAUAUAUGCAUAUAUACU ..((((.((((((........((((((((....)))))))).......))))))...))))((((((.....))))))...........(((((((........)))))))..... ( -25.56) >DroYak_CAF1 18462 96 - 1 CUGGCAUUUGCUGAUCAAUACAUGUAUGUAUGUAUGUUCAUAUUAAUACAGCAACUCUGUCAAACAGAUUCGUUGUUUUAUAUGCAUACGU-------------UGCAC------- ..(((....)))..........(((((((((((((((..........(((((...((((.....))))...)))))...))))))))))).-------------)))).------- ( -22.52) >consensus CUGGCAUGUGCUGAUCAAUACAUGUAUGUAUGU_UGUACAUAUUAAUACAGCAACACUGUCAAACAGAUUCGUUGUUUCAUAUACAUACAUAUGCA______UAUGCAUAU_UA_U ..(((....))).........(((((((((((((.............(((((....(((.....)))....)))))..)))))))))))))......................... (-14.94 = -15.73 + 0.79)


| Location | 156,250 – 156,357 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.50 |
| Mean single sequence MFE | -21.50 |
| Consensus MFE | -12.06 |
| Energy contribution | -12.07 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.613394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 156250 107 + 1281640 GCAACAAAUCUGUUU-UCAGCGUUGUUGUAUUAAUAUAU----GCAUACAUACAUGUAUUGAUCAGCACAAGCCAGCUGCACAAAAAUUUGAAUGAAUAUACAUAUGUACA----U (((((((..(((...-.)))..)))))))((((((((((----(........)))))))))))((((........)))).............(((.((((....)))).))----) ( -21.20) >DroSec_CAF1 30746 94 + 1 GCAACGAAUCUUUUUGACAGUGUUGCUGUAUUAGUAUAU----GCAUACAUACAUGUAUUGAUCAGCACAUGCCAGCUGC--AAAAAUUUUAAUGAAUA----------------U .((...((..((((((.(((((((((((.((((((((((----(........))))))))))))))))...))..)))))--)))))..))..))....----------------. ( -23.40) >DroSim_CAF1 15880 93 + 1 GCAACGAAUCUUUUUGACAGUGUUGCUGUAUUAGUAUAU----GCAUACAUACAUGUAUUG-UCAGCACAUGCCAGCUGC--AAAAAUGUUAAUGAAUA----------------U .((...(((.((((((.(((((((((((...((((((((----(........)))))))))-.)))))...))..)))))--))))).)))..))....----------------. ( -21.50) >DroYak_CAF1 18482 103 + 1 ACAACGAAUCUGUUUGACAGAGUUGCUGUAUUAAUAUGAACAUACAUACAUACAUGUAUUGAUCAGCAAAUGCCAGCUGCACAAAAAUUUGUAUGAAU-------------UACAC .(((((((((((.....))))((.((((((((....(((.(((((((......))))).)).)))...)))).)))).)).......))))).))...-------------..... ( -19.90) >consensus GCAACGAAUCUGUUUGACAGUGUUGCUGUAUUAAUAUAU____GCAUACAUACAUGUAUUGAUCAGCACAUGCCAGCUGC__AAAAAUUUGAAUGAAUA________________U .................(((((((((((.((((((((((..............)))))))))))))))...))..))))..................................... (-12.06 = -12.07 + 0.00)

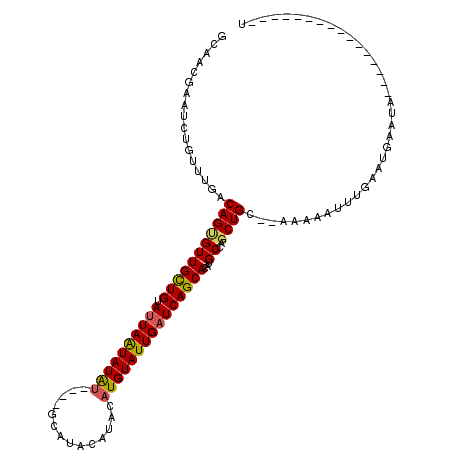
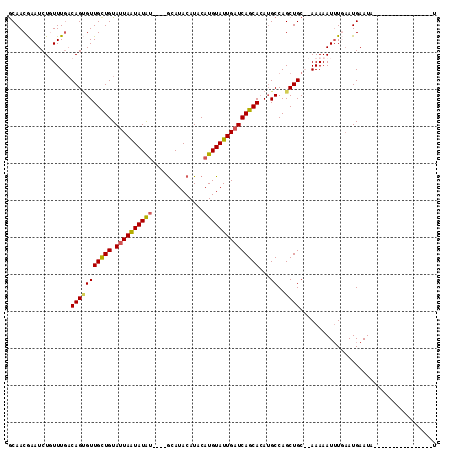
| Location | 156,288 – 156,384 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -21.33 |
| Energy contribution | -21.67 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.29 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 156288 96 + 1281640 GCAUACAUACAUGUAUUGAUCAGCACAAGCCAGCUGCACAAAAAUUUGAAUGAAUAUACAUAUGUACAUAUGUACGGUACAUGUGUAUGCAUUAAU ((((((..((((((((((..((((........))))....................(((((((....)))))))))))))))))))))))...... ( -28.80) >DroSec_CAF1 30785 82 + 1 GCAUACAUACAUGUAUUGAUCAGCACAUGCCAGCUGC--AAAAAUUUUAAUGAAUA------------UAUGUACGGUACAUGUGUAUGCAUUAAU ((((((..((((((((((..((((........)))).--....((((....)))).------------......))))))))))))))))...... ( -22.80) >DroSim_CAF1 15919 81 + 1 GCAUACAUACAUGUAUUG-UCAGCACAUGCCAGCUGC--AAAAAUGUUAAUGAAUA------------UAUGUACGGUACAUGUGUAUGCAUUAAU ((((((..((((((((((-(((((........)))).--....(((((....))))------------)....)))))))))))))))))...... ( -24.10) >consensus GCAUACAUACAUGUAUUGAUCAGCACAUGCCAGCUGC__AAAAAUUUUAAUGAAUA____________UAUGUACGGUACAUGUGUAUGCAUUAAU ((((((..((((((((((..((((........)))).......((((....))))...................))))))))))))))))...... (-21.33 = -21.67 + 0.33)

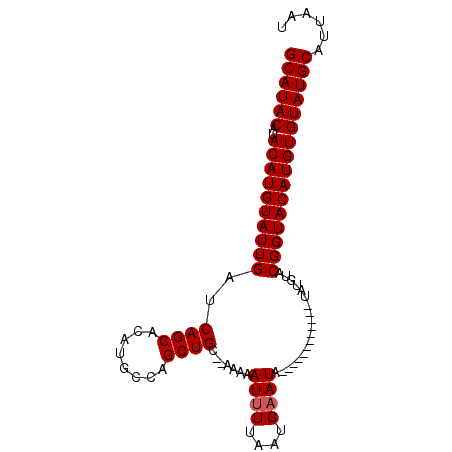
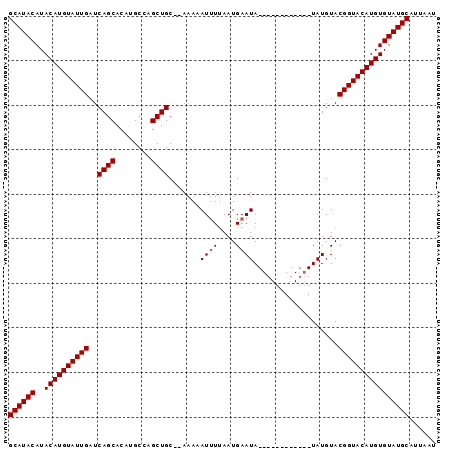
| Location | 156,288 – 156,384 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 86.86 |
| Mean single sequence MFE | -20.50 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 156288 96 - 1281640 AUUAAUGCAUACACAUGUACCGUACAUAUGUACAUAUGUAUAUUCAUUCAAAUUUUUGUGCAGCUGGCUUGUGCUGAUCAAUACAUGUAUGUAUGC ......(((((((((((((..((((((((....))))))))..............(((..((((........))))..))))))))))..)))))) ( -26.50) >DroSec_CAF1 30785 82 - 1 AUUAAUGCAUACACAUGUACCGUACAUA------------UAUUCAUUAAAAUUUUU--GCAGCUGGCAUGUGCUGAUCAAUACAUGUAUGUAUGC ......(((((((((((((.....((..------------.(((......)))...)--)((((........)))).....)))))))..)))))) ( -16.60) >DroSim_CAF1 15919 81 - 1 AUUAAUGCAUACACAUGUACCGUACAUA------------UAUUCAUUAACAUUUUU--GCAGCUGGCAUGUGCUGA-CAAUACAUGUAUGUAUGC ......(((((((.(((((...))))).------------.........((((..((--(((((........)))).-)))...)))).))))))) ( -18.40) >consensus AUUAAUGCAUACACAUGUACCGUACAUA____________UAUUCAUUAAAAUUUUU__GCAGCUGGCAUGUGCUGAUCAAUACAUGUAUGUAUGC ......(((((((((((((......(((((........))))).................((((........)))).....)))))))..)))))) (-16.00 = -16.33 + 0.33)

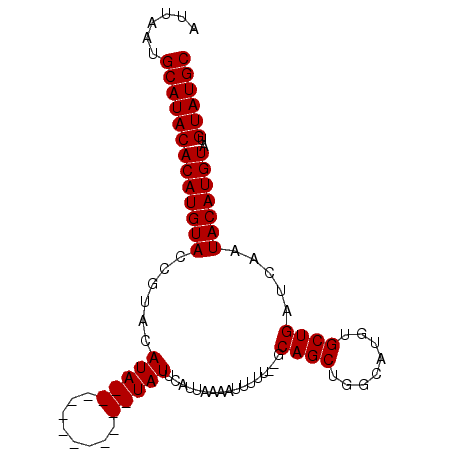
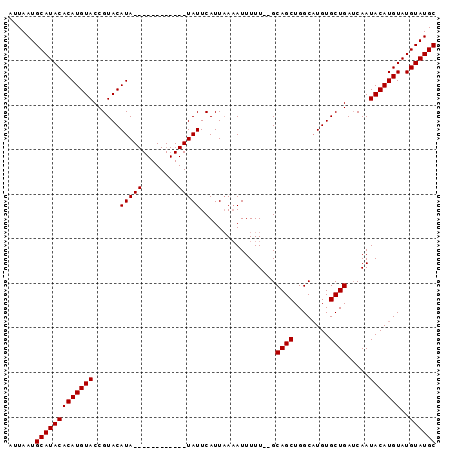
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:08 2006