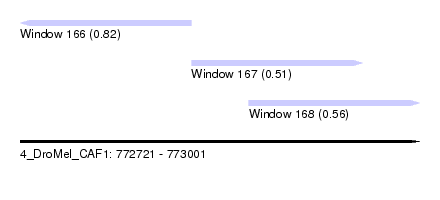
| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 772,721 – 773,001 |
| Length | 280 |
| Max. P | 0.824381 |

| Location | 772,721 – 772,841 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.78 |
| Mean single sequence MFE | -30.19 |
| Consensus MFE | -15.99 |
| Energy contribution | -16.30 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
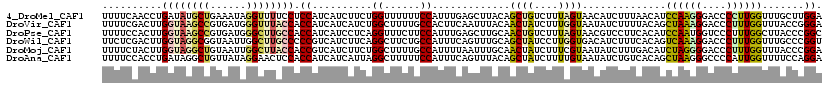
>4_DroMel_CAF1 772721 120 - 1281640 UUUUCAACCUGAUAUGCUGAAAUAGGUUUUCCUCCAUCAUCUUCUGGUUUUUUCCAUUUGAGCUUACAGCUGUCUUUAGUAACAUCUUUAACAUCCAAGGGACCCCUUGGUUUGCUUGGA .....((((.((...(.(((...(((....)))...))).).)).)))).......((..(((..((.((((....))))...............(((((....)))))))..)))..)) ( -25.00) >DroVir_CAF1 1655 120 - 1 UUUUCGACUUGGUAAGCCGUGAUGGGUUUACCACCAUCAUCAUCUGGCUUUUGCCACUUCAAUUUACAACUAUCUUUGGUAAUAUCUUUUACAGCUAAAGGACCCUUUGGUUUACCGGGA .......((((((((((((((((((........)))))))....((((....))))................((((((((..((.....))..)))))))).......))))))))))). ( -36.70) >DroPse_CAF1 1284 120 - 1 UUUUCCACUUGGUAAGCCGUGAUGGGCUUGCCACCAUCAUCCUCAGGUUUCUUCCAUUUGAGCUUGCAACUGUCUUUAGUAACGUCCUUCACAUCCAAUGGUCCCUUUGGCUUACCCGGC .......((.((((((((((((.((((..........((..(((((((.......)))))))..))..((((....))))...)))).)))).......(....)...)))))))).)). ( -32.20) >DroWil_CAF1 1770 120 - 1 UUCUCGACUUGGUAGGCGGUAAUUGGCUUGCCCCCGUCAUCUUCAGGCUUCUGCCAUUUCAGUUUGCAGCUAUCCUUGGUGACAUCUUUCACAGUCAAAGGACCCUUUGGUUUGCCCGGU ......(((.((((((((((((.....)))))...((((((...(((...((((...........))))....))).))))))............(((((....)))))))))))).))) ( -31.80) >DroMoj_CAF1 2254 120 - 1 UUUUCUACUUGGUAGGCUGUAAUUGGCUUACCACCGUCAUCUUCUGGCUUUUGCCAUUUUAAUUUGCAACUAUCUUUCGUAAUAUCUUUGACAUCUAGGGGACCCUUUGGUUUACCCGGA .........(((((((((......)))))))))((((((.....((((....)))).......((((...........))))......)))).....(((((((....))))..))))). ( -31.50) >DroAna_CAF1 3668 120 - 1 UUUUCCACCUGAUAGGCUGUUAUAGGAACUCCACCAUCAUCAUUAGGCUUUUUCCAUUUCAGUUUACAGCUAUCUUUUGUAAUAUCUGUCACAGCUAAGGGCCCCAUUGGUUUUCCAGGA .......((((..(((((......((....)).............(((((((.......(((.((((((.......))))))...)))........))))))).....)))))..)))). ( -23.96) >consensus UUUUCCACUUGGUAGGCUGUAAUAGGCUUACCACCAUCAUCUUCAGGCUUUUGCCAUUUCAGUUUACAACUAUCUUUAGUAACAUCUUUCACAGCCAAGGGACCCUUUGGUUUACCCGGA ..........((((((((......)))))))).((..........((......)).............((((....))))..............((((((....)))))).......)). (-15.99 = -16.30 + 0.31)

| Location | 772,841 – 772,961 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.44 |
| Mean single sequence MFE | -40.05 |
| Consensus MFE | -30.41 |
| Energy contribution | -31.75 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 772841 120 + 1281640 GUUUGAUAAAAAACAAGGUCGUUGGGUCCCUCUUGGUCGAACGUCUGCCAAUGAUACCGAAUUUGAUGUAAAGGGCCUCCAGGAAGGCCAUGAGUAUCAGUUUCGCGUCAAGGCAAUUAA ((((......))))..((.((((.(..(......)..).)))).))(((..((((..((((.((((((.....(((((......))))).....)))))).)))).)))).)))...... ( -35.10) >DroVir_CAF1 1775 120 + 1 AUUCGAUAAGAAACAAGGUCGAUGGGUACCUGUCGGUCGAGCCUCCGGAAAUGAUACCGAAUUUGAUGUCAAGGGCCUUCAGGAAGGCCAUGAAUAUCAGUUUAGGGUUAAAGCCAUCAA ....(((.........(..((((((....))))))..).(((((.(((........)))...(((((((((..((((((....)))))).)).)))))))....)))))......))).. ( -35.50) >DroGri_CAF1 2562 120 + 1 AUUCGAUAAGAAACAGGGUCGUUGGGUGCCGGUCGGUCGUACCUCCGGCAAUGAUACCGAACUUGAUGUCAAGGGCCUGCAGGAAGGCCAUGAGUAUCAGUUUCGGGUCAAGGCCAUCAA .(((.....)))....((((......((((((..((.....)).)))))).((((.(((((.(((((((((..(((((......))))).))).)))))).))))))))).))))..... ( -46.20) >DroWil_CAF1 1890 120 + 1 GUUCGACAAGAAACAAGGUAGAUGGGUACCUGUCGGUCGAACUGCUGGCAAUGAUACCGAAUUUGAUGUCAAGGGCCUUCAGGAAGGCCAUGAAUAUCAGUUUCGGGUCAAGGCCAUCAA (((((((..((....(((((......))))).)).)))))))((.((((..((((.(((((.(((((((((..((((((....)))))).)).))))))).)))))))))..)))).)). ( -51.20) >DroMoj_CAF1 2374 120 + 1 GUUUGAUAAGAAACAAGGUCGUUGGGUACCGGUCGGUCGUACCUCUGGCAAUGAUACUGAAUUUGAUGUCAAGGGCCUUCAGGAAGGUCAUGAGUAUCAGUUUCGGGUUAAAGCCAUCAA ..(((((..(((((...((((..((((((.((....)))))))).))))..(((((((.(.((((....))))((((((....)))))).).)))))))))))).(((....)))))))) ( -36.10) >DroAna_CAF1 3788 120 + 1 GUUUGAUAAAAAACAAGGACGAUGGGUACCUGUUGGUCGAUCUUCUGGCAAUGAUACCGAAUUUGAUGUCAAGGGUCUUCAGGAAGGCCAUGAGUAUCAGUUUCGAGUCAGAGCCAUUAA ((((......))))..(..((((((....))))))..).......((((..((((..((((.(((((((((..((((((....)))))).))).)))))).)))).))))..)))).... ( -36.20) >consensus GUUCGAUAAGAAACAAGGUCGAUGGGUACCUGUCGGUCGAACCUCUGGCAAUGAUACCGAAUUUGAUGUCAAGGGCCUUCAGGAAGGCCAUGAGUAUCAGUUUCGGGUCAAAGCCAUCAA (((((((..((....((((........)))).)).)))))))....(((..((((.(((((.(((((((((..((((((....)))))).))).)))))).)))))))))..)))..... (-30.41 = -31.75 + 1.34)

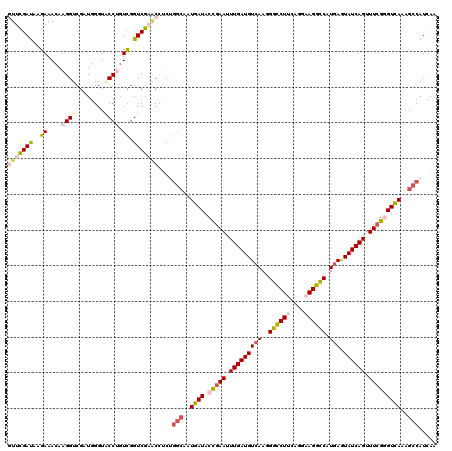
| Location | 772,881 – 773,001 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.28 |
| Mean single sequence MFE | -40.80 |
| Consensus MFE | -30.91 |
| Energy contribution | -30.50 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.559189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
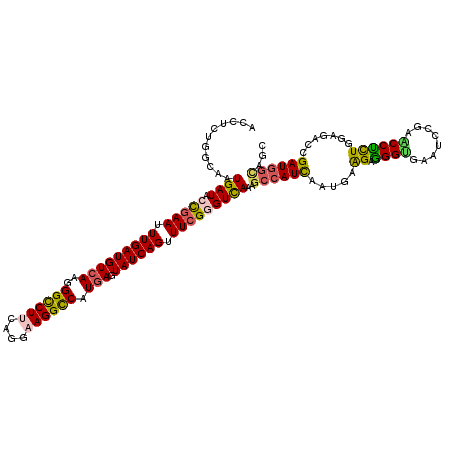
>4_DroMel_CAF1 772881 120 + 1281640 ACGUCUGCCAAUGAUACCGAAUUUGAUGUAAAGGGCCUCCAGGAAGGCCAUGAGUAUCAGUUUCGCGUCAAGGCAAUUAAUGAAGAGGGGGAAUCCGAUCCCCUGGACUCCGAUGAUAGC .((((((((..((((..((((.((((((.....(((((......))))).....)))))).)))).)))).))))...........((((((......)))))).......))))..... ( -38.40) >DroVir_CAF1 1815 120 + 1 GCCUCCGGAAAUGAUACCGAAUUUGAUGUCAAGGGCCUUCAGGAAGGCCAUGAAUAUCAGUUUAGGGUUAAAGCCAUCAAUGAGGAAGGUGAAUCCGAGCCUCUAGAGACCGAUGGAAGU .((..(((...((((.(..((((.(((((((..((((((....)))))).)).)))))))))..)(((....)))))))..((((..((.....))...))))......)))..)).... ( -36.40) >DroGri_CAF1 2602 120 + 1 ACCUCCGGCAAUGAUACCGAACUUGAUGUCAAGGGCCUGCAGGAAGGCCAUGAGUAUCAGUUUCGGGUCAAGGCCAUCAACGAGGAGGGCGAAUCCGAACCUCUGGAGACCGAUGGCAGC ..(((((((..((((.(((((.(((((((((..(((((......))))).))).)))))).)))))))))..)))......((((.(((....)))...)))).))))............ ( -46.90) >DroWil_CAF1 1930 120 + 1 ACUGCUGGCAAUGAUACCGAAUUUGAUGUCAAGGGCCUUCAGGAAGGCCAUGAAUAUCAGUUUCGGGUCAAGGCCAUCAAUGAAGAGGGUGAAUCGGAACCUCUCGAGACCGAUGGCAGC .((((((((..((((.(((((.(((((((((..((((((....)))))).)).))))))).)))))))))..)))(((......((((((........))).)))......)))))))). ( -47.90) >DroMoj_CAF1 2414 120 + 1 ACCUCUGGCAAUGAUACUGAAUUUGAUGUCAAGGGCCUUCAGGAAGGUCAUGAGUAUCAGUUUCGGGUUAAAGCCAUCAAUGAGGAAGGUGAAUCUGAACCUUUGGAGACCGAUGGUAGC ((((..(((..(((((((.(.((((....))))((((((....)))))).).))))))))))..))))....((((((......((((((........))))))(....).))))))... ( -37.00) >DroAna_CAF1 3828 120 + 1 UCUUCUGGCAAUGAUACCGAAUUUGAUGUCAAGGGUCUUCAGGAAGGCCAUGAGUAUCAGUUUCGAGUCAGAGCCAUUAAUGAAGAGGGUGAAUCUGAACCUCUUGAAACAGAUGGCAGC ...((((((........((((.(((((((((..((((((....)))))).))).)))))).)))).))))))((((((..(.((((((...........)))))).)....))))))... ( -38.20) >consensus ACCUCUGGCAAUGAUACCGAAUUUGAUGUCAAGGGCCUUCAGGAAGGCCAUGAGUAUCAGUUUCGGGUCAAAGCCAUCAAUGAAGAGGGUGAAUCCGAACCUCUGGAGACCGAUGGCAGC ...........((((.(((((.(((((((((..((((((....)))))).))).)))))).)))))))))..((((((.....((.((((........)))))).......))))))... (-30.91 = -30.50 + -0.41)

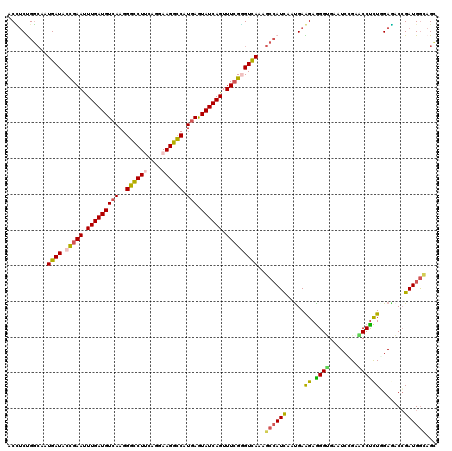
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:48 2006