| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 763,905 – 764,011 |
| Length | 106 |
| Max. P | 0.833291 |

| Location | 763,905 – 764,011 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
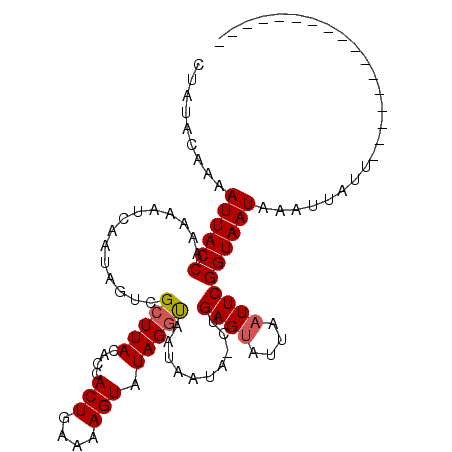
| Reading direction | forward |
| Mean pairwise identity | 84.43 |
| Mean single sequence MFE | -17.65 |
| Consensus MFE | -14.66 |
| Energy contribution | -14.06 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824261 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 763905 106 + 1281640 AUACAUAUAAAAGCAUAAAUAAUUCAUUACCGAAUUAAUACUCAG-UAUUAUUACCUAUACUUUUCAGUGGUGUAACCGACUAUUGAUUUUUGGUAAUUUUGUAUAG ....((((((((.......((((((......))))))........-.....(((((........((((((((.......)))))))).....))))))))))))).. ( -18.42) >DroSec_CAF1 20443 90 + 1 ----------------UAAUAAUUUAUUACCGAAUUAAUACUCAG-UAUUAUUGCCUAUACUUUUCAGUAGUGUAAGCGACUAUUGAAUUUUGGUAAUUUUGUACAG ----------------..((((...(((((((((.......((((-((...((((.((((((.......)))))).)))).))))))..))))))))).)))).... ( -19.50) >DroSim_CAF1 20904 90 + 1 ----------------UAAUAAUUUAUUACCGAAUUAAUACUCAG-UAUUAUUGCCUAUACUUUUCAGUAGUGUAAGCGACUAUUGAAUUUUGGUAAUUUUGUACAG ----------------..((((...(((((((((.......((((-((...((((.((((((.......)))))).)))).))))))..))))))))).)))).... ( -19.50) >DroEre_CAF1 19761 90 + 1 -----------------UAUAAUUUAUUACCGAAUUAAUACUCAGAUUUUAUUACCUAUACUUCUCAGUGGUGUAAGCGACUAUUGAUUUUUGGUAAUUUUGUAUAG -----------------((((....(((((((((((((((.((.(......((((((((........)))).)))).))).)))))))..))))))))....)))). ( -15.90) >DroYak_CAF1 18055 88 + 1 -----------------UAUAAUUUAUUACCGAAUUAAUUCUCAGAUGUGAUUACCUAUACUUUUCAGUGGUAUAAGCGACUAUUGAUUUUUGGUAAUUUUGAAU-- -----------------..((((((......))))))....(((((....((((((........((((((((.......)))))))).....)))))))))))..-- ( -14.92) >consensus _________________AAUAAUUUAUUACCGAAUUAAUACUCAG_UAUUAUUACCUAUACUUUUCAGUGGUGUAAGCGACUAUUGAUUUUUGGUAAUUUUGUAUAG ..................((((...(((((((((.(((((.........)))))..........((((((((.......))))))))..))))))))).)))).... (-14.66 = -14.06 + -0.60)

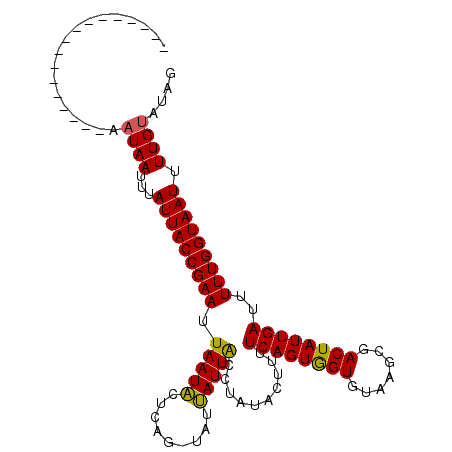
| Location | 763,905 – 764,011 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.43 |
| Mean single sequence MFE | -16.30 |
| Consensus MFE | -10.80 |
| Energy contribution | -10.96 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>4_DroMel_CAF1 763905 106 - 1281640 CUAUACAAAAUUACCAAAAAUCAAUAGUCGGUUACACCACUGAAAAGUAUAGGUAAUAAUA-CUGAGUAUUAAUUCGGUAAUGAAUUAUUUAUGCUUUUAUAUGUAU ..(((((.................((((.((.....))))))(((((((((((((((..((-((((((....))))))))....)))))))))))))))...))))) ( -25.20) >DroSec_CAF1 20443 90 - 1 CUGUACAAAAUUACCAAAAUUCAAUAGUCGCUUACACUACUGAAAAGUAUAGGCAAUAAUA-CUGAGUAUUAAUUCGGUAAUAAAUUAUUA---------------- .........((((((..............(((((...((((....)))))))))..(((((-(...))))))....)))))).........---------------- ( -16.50) >DroSim_CAF1 20904 90 - 1 CUGUACAAAAUUACCAAAAUUCAAUAGUCGCUUACACUACUGAAAAGUAUAGGCAAUAAUA-CUGAGUAUUAAUUCGGUAAUAAAUUAUUA---------------- .........((((((..............(((((...((((....)))))))))..(((((-(...))))))....)))))).........---------------- ( -16.50) >DroEre_CAF1 19761 90 - 1 CUAUACAAAAUUACCAAAAAUCAAUAGUCGCUUACACCACUGAGAAGUAUAGGUAAUAAAAUCUGAGUAUUAAUUCGGUAAUAAAUUAUA----------------- .........((((((...(((.((((.((.........(((....)))..((((......)))))).)))).))).))))))........----------------- ( -10.50) >DroYak_CAF1 18055 88 - 1 --AUUCAAAAUUACCAAAAAUCAAUAGUCGCUUAUACCACUGAAAAGUAUAGGUAAUCACAUCUGAGAAUUAAUUCGGUAAUAAAUUAUA----------------- --.......((((((..............((((((((.........))))))))............(((....)))))))))........----------------- ( -12.80) >consensus CUAUACAAAAUUACCAAAAAUCAAUAGUCGCUUACACCACUGAAAAGUAUAGGUAAUAAUA_CUGAGUAUUAAUUCGGUAAUAAAUUAUU_________________ .........((((((..............(((((....(((....))).)))))..........((((....))))))))))......................... (-10.80 = -10.96 + 0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:43 2006