| Sequence ID | 4_DroMel_CAF1 |
|---|---|
| Location | 762,711 – 762,820 |
| Length | 109 |
| Max. P | 0.891656 |

| Location | 762,711 – 762,820 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 75.24 |
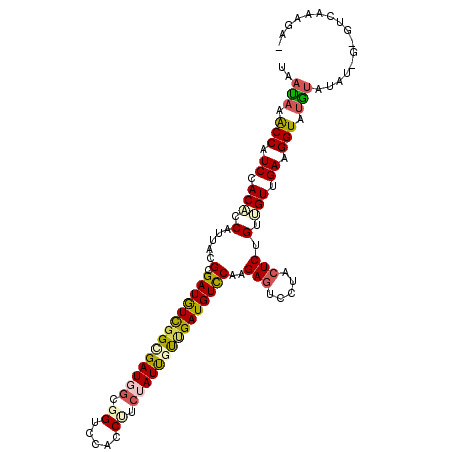
| Mean single sequence MFE | -29.92 |
| Consensus MFE | -19.92 |
| Energy contribution | -20.07 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891656 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
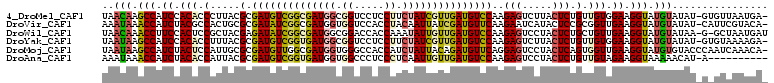
>4_DroMel_CAF1 762711 109 + 1281640 UAACAAGCCAUCCACACCCUUACGCGAUGUCGGCGAUGGCGGUCCUCCUUCUAUCGUUGAUGUCCAAGAGUCUUACUCUGUUGUGGAAGGUAUGUAUAU-GUGUUAAUGA- ..(((.(((.((((((.......(.((((((((((((((.((....))..))))))))))))))).((((.....))))..)))))).))).)))....-..........- ( -38.10) >DroVir_CAF1 21559 109 + 1 AAAUAAACCAUCUACGCCACUGCGCGAUAUCGGCGAUGGUGGUCCACCUACAAUUAUCGAUGUUCAAGAAUCAUACUCCGCGGUUGAAGGUAUGUAUAU-CAUUCGUACA- ......((((((..((((.............))))..))))))..((((.((((..(.((.((...........)))).)..)))).)))).(((((..-.....)))))- ( -21.72) >DroWil_CAF1 22898 109 + 1 UAACAAACCUUCCACUCCGCUACGAGAUAUCGGCGAUGGCGGACCACCAAAUAUUGUUGAUGUCCAAGAGUCCUACUCUGCUGUUGAAGGUAUGUAUAA-G-GCUAAUGAU ..(((.((((((.((...((...(.(((((((((((((..((....))...)))))))))))))).((((.....)))))).)).)))))).)))....-.-......... ( -32.40) >DroYak_CAF1 16708 109 + 1 UAAUAAGCCAUCCACACCUUUACGCGAUGUCGGUGAUGGCGGUCCUCCUUCUAUCGUUGAUGUCCAAGAGUCUUACUCUGUUGUGGAAGGUAUGUAUAU-GUGUAAAAGA- ..(((.(((.((((((.......(.((((((..((((((.((....))..))))))..))))))).((((.....))))..)))))).))).)))....-..........- ( -32.40) >DroMoj_CAF1 24782 110 + 1 UAAUAAGCCAUCUACUCCAUUGCGCGAUGUUGGCGAUGGUGGGCCACCAUCUAUUACAGAUGUUCAGGAGUCCUACUCAGUGGUUGAAGGUAUGUGUACCCAAUCAAACA- ..(((.(((.((..(.(((((((.((....))))))))).)(((((((((((.....))))).....(((.....))).)))))))).))).)))...............- ( -28.60) >DroAna_CAF1 326467 100 + 1 AAAUAAACCAUCUACACCAUUACGCGAUGUCGGUGAUGGUGGCCCUCCCUCAAUUGUUGAUGUCCAAGAGUCCUACUCUGUUGUAGAAGGUAAAAACAU-A---------- ......(((.((((((.(.....(.((((((((..((.(.((.....)).).))..))))))))).((((.....))))).)))))).)))........-.---------- ( -26.30) >consensus UAAUAAACCAUCCACACCAUUACGCGAUGUCGGCGAUGGCGGUCCACCUUCUAUUGUUGAUGUCCAAGAGUCCUACUCUGUUGUUGAAGGUAUGUAUAU_G_GUCAAAGA_ ..(((.(((.((.(((.(.....(.((((((((((((((.((.....)).)))))))))))))))..(((.....))).).))).)).))).)))................ (-19.92 = -20.07 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:41 2006