
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,773,080 – 26,773,235 |
| Length | 155 |
| Max. P | 0.999846 |

| Location | 26,773,080 – 26,773,195 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -23.27 |
| Energy contribution | -22.91 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26773080 115 + 27905053 -----CCUAGUUCUAUUUAUUCCAUAUUUGCAGAGGCAUUGAUAACCGAAUCAGAAAAGCUUUGAGCAAGUGGCCAAAUAUGCAAAGCUUAACUGAUUUGGAAUGCAGCUUAAUCCACAU -----..........................((..(((((.....(((((((((..(((((((..(((.((......)).))))))))))..))))))))))))))..)).......... ( -26.40) >DroSec_CAF1 76497 115 + 1 -----CCUAGCUCUUUUUAUUCCAUAUUUGCCGAGGCAUUAAUAACCGAAUCAGAAAAGCUUUGAGAAAUUGGCCAAAUAUGCAAAGCUUAACUGAUUUGGAAUGCAGCUUAAUCCACAU -----...((((................(((....))).......(((((((((..((((((((....(((.....)))...))))))))..))))))))).....)))).......... ( -27.80) >DroSim_CAF1 80250 115 + 1 -----CCUAGCUCUUUUUAUUCCAUAUUUGCCGAGGCAUUGAUAACCGAAUCAGAAAAGCUUUGAGAAAUUGGCCAAAUAUGCAAAGCUUAACUGAUUUGGAAUGCAGCUUAAUCCACAU -----...((((............(((((((....)))..)))).(((((((((..((((((((....(((.....)))...))))))))..))))))))).....)))).......... ( -28.00) >DroEre_CAF1 78010 120 + 1 CUGUCCCCAACUCCCUUUAUUCCAUAUUAGCCGAGGCAUUGAUAACCAAAUCAGAAAAGCUUUGAGCAUGUGGCCAAAUAUGCAAAGCUUAACUGAUUGGGAAUGCAGUUUAAUCCGCUU ((((.(((................(((((((....))..)))))....((((((..(((((((..((((((......)))))))))))))..)))))))))...))))............ ( -29.30) >DroYak_CAF1 79564 94 + 1 --------AA------------------UGCCGAGGCAUUGAUAACCGAAUCGGAAAAGCUUUGAGCAAGUGGCCAAAUAUGCAGAGCUUAACUGAUUUGGAAUGCAGUUUAAUCCGCUU --------((------------------(((....))))).....(((((((((..(((((((..(((.((......)).))))))))))..)))))))))...((.(......).)).. ( -27.00) >consensus _____CCUAGCUCUUUUUAUUCCAUAUUUGCCGAGGCAUUGAUAACCGAAUCAGAAAAGCUUUGAGCAAGUGGCCAAAUAUGCAAAGCUUAACUGAUUUGGAAUGCAGCUUAAUCCACAU .............................((....(((((.....(((((((((..((((((((..................))))))))..)))))))))))))).))........... (-23.27 = -22.91 + -0.36)

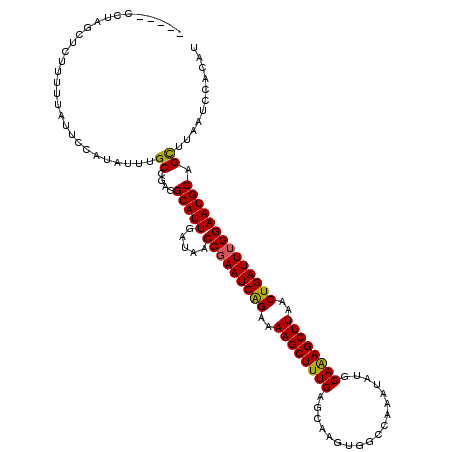
| Location | 26,773,115 – 26,773,235 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -32.05 |
| Energy contribution | -31.29 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.91 |
| SVM decision value | 4.24 |
| SVM RNA-class probability | 0.999846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26773115 120 + 27905053 GAUAACCGAAUCAGAAAAGCUUUGAGCAAGUGGCCAAAUAUGCAAAGCUUAACUGAUUUGGAAUGCAGCUUAAUCCACAUGAUACGAGCUCGUUGAGCUGAUGUUUCUCGAAUGCAGGAA .....(((((((((..(((((((..(((.((......)).))))))))))..)))))))))..((((((((.(((.....)))..))))(((..((((....))))..))).)))).... ( -34.90) >DroSec_CAF1 76532 120 + 1 AAUAACCGAAUCAGAAAAGCUUUGAGAAAUUGGCCAAAUAUGCAAAGCUUAACUGAUUUGGAAUGCAGCUUAAUCCACAUGAUACGGGCUCGUUGAGCUGAUGUUUCUCGAAUGCAGUAA .....(((((((((..((((((((....(((.....)))...))))))))..)))))))))..((((((((.(((.....)))..))))(((..((((....))))..))).)))).... ( -33.80) >DroSim_CAF1 80285 120 + 1 GAUAACCGAAUCAGAAAAGCUUUGAGAAAUUGGCCAAAUAUGCAAAGCUUAACUGAUUUGGAAUGCAGCUUAAUCCACAUGAUACGGGCUCGUUGAGCUGAUGUUCCUCGAAUGCAGGAA .....(((((((((..((((((((....(((.....)))...))))))))..)))))))))..((((((((.(((.....)))..))))(((..((((....))))..))).)))).... ( -35.50) >DroEre_CAF1 78050 120 + 1 GAUAACCAAAUCAGAAAAGCUUUGAGCAUGUGGCCAAAUAUGCAAAGCUUAACUGAUUGGGAAUGCAGUUUAAUCCGCUUGAUACGAGCUCGAUGAGCUGAUGUUUCUCGAAUGCAGGAA .....((.((((((..(((((((..((((((......)))))))))))))..))))))(((((..(((((((.((.(((((...)))))..)))))))))....))))).......)).. ( -37.80) >DroYak_CAF1 79578 120 + 1 GAUAACCGAAUCGGAAAAGCUUUGAGCAAGUGGCCAAAUAUGCAGAGCUUAACUGAUUUGGAAUGCAGUUUAAUCCGCUUGAUACGAGCUCGAUGAGCUGAUGUUUCCCGAAUGCAGAAA .....(((((((((..(((((((..(((.((......)).))))))))))..)))))))))..((((.........(((((...)))))(((..((((....))))..))).)))).... ( -34.40) >consensus GAUAACCGAAUCAGAAAAGCUUUGAGCAAGUGGCCAAAUAUGCAAAGCUUAACUGAUUUGGAAUGCAGCUUAAUCCACAUGAUACGAGCUCGUUGAGCUGAUGUUUCUCGAAUGCAGGAA .....(((((((((..((((((((..................))))))))..)))))))))..((((((((.(((.....)))..))))(((..((((....))))..))).)))).... (-32.05 = -31.29 + -0.76)



| Location | 26,773,115 – 26,773,235 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.25 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -23.29 |
| Energy contribution | -23.69 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619024 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26773115 120 - 27905053 UUCCUGCAUUCGAGAAACAUCAGCUCAACGAGCUCGUAUCAUGUGGAUUAAGCUGCAUUCCAAAUCAGUUAAGCUUUGCAUAUUUGGCCACUUGCUCAAAGCUUUUCUGAUUCGGUUAUC .(((.((((.(((((....)).((((...)))))))....)))))))............((.((((((..((((((((.......(((.....)))))))))))..)))))).))..... ( -28.41) >DroSec_CAF1 76532 120 - 1 UUACUGCAUUCGAGAAACAUCAGCUCAACGAGCCCGUAUCAUGUGGAUUAAGCUGCAUUCCAAAUCAGUUAAGCUUUGCAUAUUUGGCCAAUUUCUCAAAGCUUUUCUGAUUCGGUUAUU ...(..(((..((......)).((((...)))).......)))..).............((.((((((..((((((((.....((....)).....))))))))..)))))).))..... ( -25.40) >DroSim_CAF1 80285 120 - 1 UUCCUGCAUUCGAGGAACAUCAGCUCAACGAGCCCGUAUCAUGUGGAUUAAGCUGCAUUCCAAAUCAGUUAAGCUUUGCAUAUUUGGCCAAUUUCUCAAAGCUUUUCUGAUUCGGUUAUC (((((.......))))).....((((...)))).......(((..(......)..))).((.((((((..((((((((.....((....)).....))))))))..)))))).))..... ( -27.70) >DroEre_CAF1 78050 120 - 1 UUCCUGCAUUCGAGAAACAUCAGCUCAUCGAGCUCGUAUCAAGCGGAUUAAACUGCAUUCCCAAUCAGUUAAGCUUUGCAUAUUUGGCCACAUGCUCAAAGCUUUUCUGAUUUGGUUAUC .............((.((...(((((...))))).)).))..((((......))))...((.((((((..((((((((.......(((.....)))))))))))..)))))).))..... ( -29.41) >DroYak_CAF1 79578 120 - 1 UUUCUGCAUUCGGGAAACAUCAGCUCAUCGAGCUCGUAUCAAGCGGAUUAAACUGCAUUCCAAAUCAGUUAAGCUCUGCAUAUUUGGCCACUUGCUCAAAGCUUUUCCGAUUCGGUUAUC ...(((...(((((((.....((((....((((..((.....((((......))))...(((((((((.......)))...))))))..))..))))..)))))))))))..)))..... ( -27.50) >consensus UUCCUGCAUUCGAGAAACAUCAGCUCAACGAGCUCGUAUCAUGUGGAUUAAGCUGCAUUCCAAAUCAGUUAAGCUUUGCAUAUUUGGCCACUUGCUCAAAGCUUUUCUGAUUCGGUUAUC ....(((....((......)).((((...))))..)))....((((......))))...((.((((((..((((((((.......(((.....)))))))))))..)))))).))..... (-23.29 = -23.69 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:45 2006