
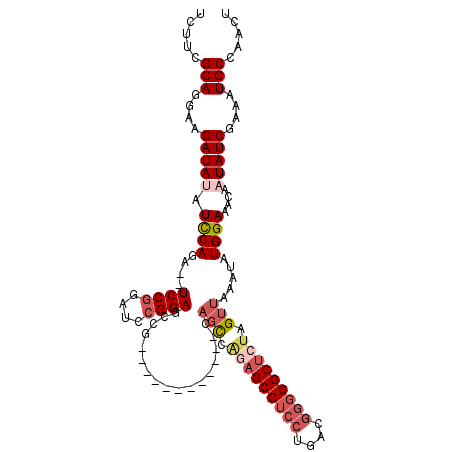
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,764,734 – 26,764,835 |
| Length | 101 |
| Max. P | 0.952888 |

| Location | 26,764,734 – 26,764,835 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 85.31 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -24.60 |
| Energy contribution | -26.56 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.952888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26764734 101 + 27905053 UCUUCGCAGGAAGAUAUAUCCAGA--UCCGGAUCCGGAGCCG------------GAGUCAGAGCCUCCUGACGGGGGCUCUAGUUAAAUAUGGAAACAAUAUCGAAAUGCCAACU .....(((....(((((.(((.(.--((((....)))).).)------------))...(((((((((....))))))))).........((....)))))))....)))..... ( -36.40) >DroSec_CAF1 68525 101 + 1 UCUUCGCAGGAAGAUAUAUCCAGA--UCCGGAUCCGGAGCCG------------GAGCCAGAGCCUCCUGACUGGGGCUCUAGUUAAAUAUGGAAACAAUAUCGAAAUGCCAACU .....(((....(((((.(((.(.--((((....)))).).)------------))((.((((((.((.....)))))))).))......((....)))))))....)))..... ( -33.70) >DroSim_CAF1 72200 100 + 1 UCUUCGCAGGAAGAUAUAUCCAGA--UCCGGAUCCGGAGCCG-------------AGCCAGAGCCUCCUGACGGGGGCUCUAGUUAAAUAUGGAAACAAUAUCGAAAUGCCAACU .....(((....(((((.((((..--((((....))))....-------------(((.(((((((((....))))))))).))).....))))....)))))....)))..... ( -35.40) >DroEre_CAF1 69821 99 + 1 UCUUCGCAGAAAGAUACAUCCAGA--UCCGGAUCCGGAUCCU-----------GC--CUCCUGCCCACUGACGG-GGCUCUAGUUAAAUAUGGAAACAAUAUCGAAAUGCCAGCU .....(((....((((..((((((--((((....))))))..-----------..--.....((((.......)-)))............)))).....))))....)))..... ( -25.90) >DroYak_CAF1 71184 115 + 1 UCUUCGCAGGAAGAUACAUUCAGAUAUCCAGAUCCGGAUGCAGAAAUGGAGCUGCAGCUGAAGCCUCCUGACGGGGGCUCUAGUUAAAUAUGGAAACAAUAUCGAAAUGCCAACU (((((....)))))..((((..(((((.....((((...((((........))))(((((.(((((((....))))))).))))).....))))....)))))..))))...... ( -34.50) >consensus UCUUCGCAGGAAGAUAUAUCCAGA__UCCGGAUCCGGAGCCG____________CAGCCAGAGCCUCCUGACGGGGGCUCUAGUUAAAUAUGGAAACAAUAUCGAAAUGCCAACU .....(((....(((((.((((....((((....)))).................(((.(((((((((....))))))))).))).....))))....)))))....)))..... (-24.60 = -26.56 + 1.96)


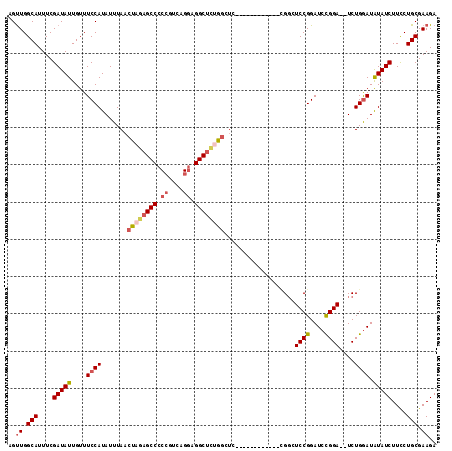
| Location | 26,764,734 – 26,764,835 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 85.31 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -24.94 |
| Energy contribution | -25.98 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26764734 101 - 27905053 AGUUGGCAUUUCGAUAUUGUUUCCAUAUUUAACUAGAGCCCCCGUCAGGAGGCUCUGACUC------------CGGCUCCGGAUCCGGA--UCUGGAUAUAUCUUCCUGCGAAGA ..((.(((....(((((................(((((((.((....)).)))))))..((------------(((.((((....))))--.))))).)))))....))).)).. ( -34.00) >DroSec_CAF1 68525 101 - 1 AGUUGGCAUUUCGAUAUUGUUUCCAUAUUUAACUAGAGCCCCAGUCAGGAGGCUCUGGCUC------------CGGCUCCGGAUCCGGA--UCUGGAUAUAUCUUCCUGCGAAGA ..((.(((....(((((...............((((((((((.....)).)))))))).((------------(((.((((....))))--.))))).)))))....))).)).. ( -35.70) >DroSim_CAF1 72200 100 - 1 AGUUGGCAUUUCGAUAUUGUUUCCAUAUUUAACUAGAGCCCCCGUCAGGAGGCUCUGGCU-------------CGGCUCCGGAUCCGGA--UCUGGAUAUAUCUUCCUGCGAAGA ..((.(((....(((((....(((........((((((((.((....)).))))))))..-------------.((.((((....))))--.))))).)))))....))).)).. ( -34.10) >DroEre_CAF1 69821 99 - 1 AGCUGGCAUUUCGAUAUUGUUUCCAUAUUUAACUAGAGCC-CCGUCAGUGGGCAGGAG--GC-----------AGGAUCCGGAUCCGGA--UCUGGAUGUAUCUUUCUGCGAAGA ..((.(((....((((((((((((...(((....)))(((-((....).)))).))))--))-----------)(((((((....))))--)))....)))))....)))..)). ( -31.80) >DroYak_CAF1 71184 115 - 1 AGUUGGCAUUUCGAUAUUGUUUCCAUAUUUAACUAGAGCCCCCGUCAGGAGGCUUCAGCUGCAGCUCCAUUUCUGCAUCCGGAUCUGGAUAUCUGAAUGUAUCUUCCUGCGAAGA .....((((((.((((((...(((........((..((((.((....)).))))..)).(((((........)))))...)))....)))))).)))))).(((((....))))) ( -33.40) >consensus AGUUGGCAUUUCGAUAUUGUUUCCAUAUUUAACUAGAGCCCCCGUCAGGAGGCUCUGGCUC____________CGGCUCCGGAUCCGGA__UCUGGAUAUAUCUUCCUGCGAAGA ..((.(((....(((((....((((.......((((((((.((....)).))))))))...................((((....))))....)))).)))))....))).)).. (-24.94 = -25.98 + 1.04)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:32 2006