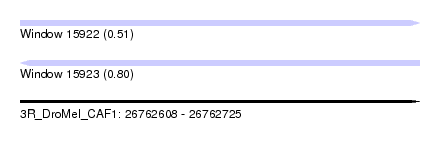
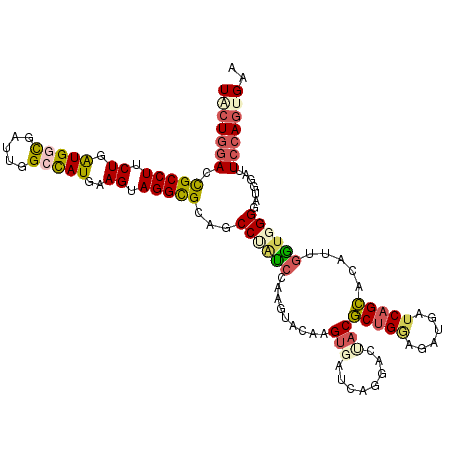
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,762,608 – 26,762,725 |
| Length | 117 |
| Max. P | 0.801173 |

| Location | 26,762,608 – 26,762,725 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -40.32 |
| Consensus MFE | -27.36 |
| Energy contribution | -28.12 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514491 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26762608 117 + 27905053 UACUGGACCGCCUUCUGAUGGCGAUUGGCCAUGAAGUAGGCGCAGCCUAUCCAAAUACAAGUGAUCAGGACUACGCUGGAGAUGAUCAGCACUUUGGUGGGUAUGGAUUCCAGUGAA (((((((((((((.((.(((((.....)))))..)).))))...(((((.(((((.....(((........)))(((((......)))))..))))))))))..))..))))))).. ( -42.00) >DroSec_CAF1 66468 117 + 1 UACUGGACCGCCUUCUGAUGGUGAUUGGCCAUGAAGUAGGCGCAGCCAAUCCAAGUACAAGUGAUCAGGACUACGCUGGAGAUGAUCAGCACAUUUGUGGGGAUGGAUUCCAGUGAA (((((((.(((((.((.(((((.....)))))..)).)))))...(((.(((...((((((((....(.....)(((((......))))).)))))))).))))))..))))))).. ( -42.20) >DroSim_CAF1 70050 117 + 1 UACUGGACCGCCUUCUGAUGGCGAUUGGCCAUGAAGUAGGCGCAGCCAAUCCAAGUACAAGUGAUCAGGACUACGCUGGAGAUGAUCAGCACAUUGGUGGGGAUGGAUUCCAGUGAA (((((((.(((((.((.(((((.....)))))..)).)))))...(((.(((...(((.((((....(.....)(((((......))))).)))).))).))))))..))))))).. ( -40.30) >DroEre_CAF1 67689 117 + 1 UGCUGGACCGCCUUCUGAUGCCGAUUGGCCAUGAAGUAGGCGCAGCCUGUCCAAGUGAGAGUGAUCAGGACCACGCUGGCGAAGAUCAGCACUUUGGUGGGGAUGGAUUCCAGUGUG .((((((.(((((.((.(((((....)).)))..)).)))))((.((..((.(((((....(((((....((.....))....))))).))))).))..))..))...))))))... ( -39.00) >DroYak_CAF1 69059 117 + 1 UACUGGACCGCCUUCUGGUGGCGAUUGGCCAUGAAGAAGGCGCAGCCUACCCAAGUGACCGUGAUCACGACGACGCUGCAGAGGAUCAGCACCUUGGUGGGUACGGAUUCCAGCGUA ..((((((((((((((.(((((.....)))))..)))))))...(((((((...((((......))))......((((........)))).....)))))))..))..))))).... ( -47.20) >DroPer_CAF1 76294 116 + 1 UACUGCAGUGCCUUCUGGUGCCUGCUGGUUAUGAAGUAGGUGCAGCCCAGCCAGACGGUGGACAAUACGAUAGCUCCGAUAAUUAUCAGAACGUUGCGGGGUAUUUCUUCGAU-GAA ..((((...(((....)))(((((((........))))))))))).(((.((....)))))......(((..((((((.((((.........))))))))))......)))..-... ( -31.20) >consensus UACUGGACCGCCUUCUGAUGGCGAUUGGCCAUGAAGUAGGCGCAGCCUAUCCAAGUACAAGUGAUCAGGACUACGCUGGAGAUGAUCAGCACAUUGGUGGGGAUGGAUUCCAGUGAA (((((((.(((((.((.(((((.....)))))..)).)))))...((((((.........(((........)))(((((......))))).....)))))).......))))))).. (-27.36 = -28.12 + 0.76)


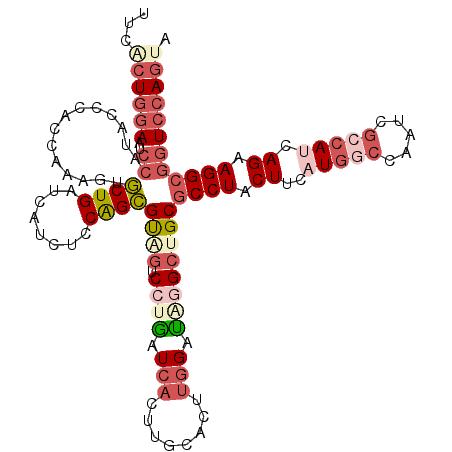
| Location | 26,762,608 – 26,762,725 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -21.41 |
| Energy contribution | -24.17 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26762608 117 - 27905053 UUCACUGGAAUCCAUACCCACCAAAGUGCUGAUCAUCUCCAGCGUAGUCCUGAUCACUUGUAUUUGGAUAGGCUGCGCCUACUUCAUGGCCAAUCGCCAUCAGAAGGCGGUCCAGUA ...((((((..((..............((((........))))((((.((((.(((........))).))))))))((((.((..(((((.....))))).)).)))))))))))). ( -38.50) >DroSec_CAF1 66468 117 - 1 UUCACUGGAAUCCAUCCCCACAAAUGUGCUGAUCAUCUCCAGCGUAGUCCUGAUCACUUGUACUUGGAUUGGCUGCGCCUACUUCAUGGCCAAUCACCAUCAGAAGGCGGUCCAGUA ...((((((..((..............((((........))))((((((..((((.(........)))))))))))((((.((..((((.......)))).)).)))))))))))). ( -33.90) >DroSim_CAF1 70050 117 - 1 UUCACUGGAAUCCAUCCCCACCAAUGUGCUGAUCAUCUCCAGCGUAGUCCUGAUCACUUGUACUUGGAUUGGCUGCGCCUACUUCAUGGCCAAUCGCCAUCAGAAGGCGGUCCAGUA ...((((((..((..............((((........))))((((((..((((.(........)))))))))))((((.((..(((((.....))))).)).)))))))))))). ( -37.40) >DroEre_CAF1 67689 117 - 1 CACACUGGAAUCCAUCCCCACCAAAGUGCUGAUCUUCGCCAGCGUGGUCCUGAUCACUCUCACUUGGACAGGCUGCGCCUACUUCAUGGCCAAUCGGCAUCAGAAGGCGGUCCAGCA (((.((((.....(((..(((....)))..))).....)))).)))(((((((......)))...))))..((((.(((..((((...(((....)))....))))..))).)))). ( -38.70) >DroYak_CAF1 69059 117 - 1 UACGCUGGAAUCCGUACCCACCAAGGUGCUGAUCCUCUGCAGCGUCGUCGUGAUCACGGUCACUUGGGUAGGCUGCGCCUUCUUCAUGGCCAAUCGCCACCAGAAGGCGGUCCAGUA ...((((((..((.((((((....(..((((........))))..)...(((((....))))).))))))))...((((((((...((((.....))))..)))))))).)))))). ( -50.50) >DroPer_CAF1 76294 116 - 1 UUC-AUCGAAGAAAUACCCCGCAACGUUCUGAUAAUUAUCGGAGCUAUCGUAUUGUCCACCGUCUGGCUGGGCUGCACCUACUUCAUAACCAGCAGGCACCAGAAGGCACUGCAGUA ...-...((((..............((((((((....))))))))....(((..(((((((....)).)))))))).....)))).......((((((........)).)))).... ( -30.30) >consensus UUCACUGGAAUCCAUACCCACCAAAGUGCUGAUCAUCUCCAGCGUAGUCCUGAUCACUUGCACUUGGAUAGGCUGCGCCUACUUCAUGGCCAAUCGCCAUCAGAAGGCGGUCCAGUA ...((((((..((..............((((........))))((((.((((.(((........))).))))))))((((.((..(((((.....))))).)).)))))))))))). (-21.41 = -24.17 + 2.75)

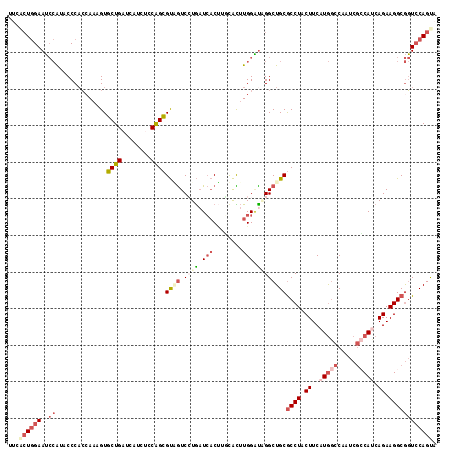
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:30 2006