
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,743,815 – 26,743,938 |
| Length | 123 |
| Max. P | 0.942432 |

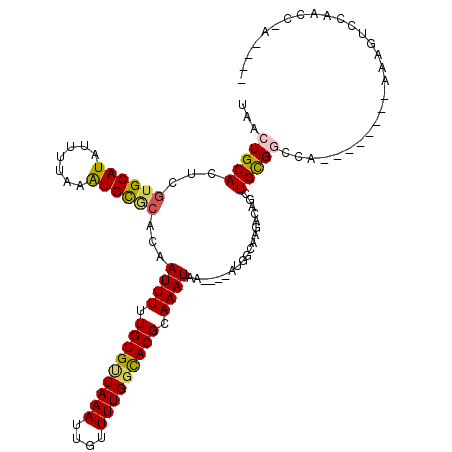
| Location | 26,743,815 – 26,743,915 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.38 |
| Mean single sequence MFE | -27.42 |
| Consensus MFE | -17.48 |
| Energy contribution | -17.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26743815 100 - 27905053 UAACUGCAGUCGUGCAUAUUUUAAAUGCCAGCCAUUUUUGUGCUAAAUUGUUUUGGCACGCAAAUAA----AUGGCAGGACAUCUGUGGCCA--------UGGGUUCACCC-A---- ...(..(((..((((((.......))))..(((((((.(((((((((....))))))))).....))----)))))...))..)))..)...--------((((....)))-)---- ( -32.90) >DroVir_CAF1 60697 103 - 1 UAACUGCACUCGUGCAUAUUUUAAGUGCGCACAAUUUUUGUGUUAAAUUGUUUUGCCACGCAAAUAA----AUGGCCAGACACCUGCGGCAA--------CCAUACCAAACCACC-- ...(((((...((((((.......))))))..................(((((.((((.........----.)))).)))))..)))))...--------...............-- ( -20.50) >DroGri_CAF1 70426 109 - 1 UAGCUGCACUCGUGCAUAUUUUAAAUGCGCACAAUUUUUGUGUUAAAUUGUUUUGGUACGCAAAUAAAAUAAUGGCCAGACAGCUGCGGCUA--------AAAUGGCAACACACCUU ((((((((((.((((((.......))))))(((((((.......)))))))((((((.................)))))).)).))))))))--------...((....))...... ( -26.73) >DroWil_CAF1 55462 100 - 1 UAACUGCAGUCGUGCAUAUUUUAAAUGUGCACAAUUUUUGUGUUAAAUUGUUUUGGCACGCAAAUAA----AUAUCAGAACAGCUGCGUCCC--------AAUGUACACAC-A---- .....(((((.((((((((.....)))))))).((((.(((((((((....))))))))).))))..----...........))))).....--------...........-.---- ( -25.80) >DroMoj_CAF1 51694 101 - 1 UAACUGCACUCGUGCAUAUUUUAAAUGCGCACAAUUUUUGUGUUAAAUUGUUUUGGCACGCAAAUAA----AUGGCCAGGCAGCUGCGACCA--------CAAUGCCAACC-GC--- ....(((....((((((.......)))))).........((((((((....))))))))))).....----..((...((((..((......--------)).))))..))-..--- ( -24.90) >DroAna_CAF1 56783 108 - 1 UAACUGCAGUCGUGCAUAUUUUAAAUGCACACCAUUUUUGUGCUAAAUUGUUUUGGCACGCAAAUAA----AUGGCAGGACAUCUGUGGCCAGGACACACUGGGUUCACCG-C---- .....((.(((((((((.......)))))).((((((.(((((((((....))))))))).....))----))))...)))....((((((((......))))..)))).)-)---- ( -33.70) >consensus UAACUGCACUCGUGCAUAUUUUAAAUGCGCACAAUUUUUGUGUUAAAUUGUUUUGGCACGCAAAUAA____AUGGCAAGACAGCUGCGGCCA________AAAGUCCAACC_A____ ...(((((...((((((.......))))))...((((.(((((((((....))))))))).))))...................)))))............................ (-17.48 = -17.40 + -0.08)


| Location | 26,743,839 – 26,743,938 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.83 |
| Mean single sequence MFE | -29.93 |
| Consensus MFE | -18.47 |
| Energy contribution | -19.25 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651295 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26743839 99 - 27905053 --GUGUUUG------UA------CG---GGUGUCAAAGGAUAACUGCAGUCGUGCAUAUUUUAAAUGCCAGCCAUUUUUGUGCUAAAUUGUUUUGGCACGCAAAUAAAUGGCAGGA --.....((------((------((---(.(((...((.....))))).)))))))...........((.(((((((.(((((((((....))))))))).....))))))).)). ( -31.70) >DroVir_CAF1 60724 107 - 1 GAGUGUGUG------UCCUGGUGUU---CAUGUCAAAGGAUAACUGCACUCGUGCAUAUUUUAAGUGCGCACAAUUUUUGUGUUAAAUUGUUUUGCCACGCAAAUAAAUGGCCAGA ((((((.((------((((..((..---.....)).))))))...))))))((((((.......))))))(((((((.......)))))))...((((..........)))).... ( -29.50) >DroWil_CAF1 55486 97 - 1 --GUGUUUG------UC------C-----GUGUCAAAGGAUAACUGCAGUCGUGCAUAUUUUAAAUGUGCACAAUUUUUGUGUUAAAUUGUUUUGGCACGCAAAUAAAUAUCAGAA --(((((((------(.------(-----(((((((((.(((((.((((..((((((((.....)))))))).....))))))))...).))))))))))...))))))))..... ( -27.10) >DroMoj_CAF1 51719 107 - 1 GUGUGUGUG------UCCUGGUGUG---CAUGUCAAAGGAUAACUGCACUCGUGCAUAUUUUAAAUGCGCACAAUUUUUGUGUUAAAUUGUUUUGGCACGCAAAUAAAUGGCCAGG .........------.(((((((((---((((((....))))..)))))..((((((.......))))))...((((.(((((((((....))))))))).)))).....)))))) ( -32.60) >DroAna_CAF1 56815 108 - 1 --CUUUUUCGGGGCGUG------UGAGUUGUGUCAAAGGAUAACUGCAGUCGUGCAUAUUUUAAAUGCACACCAUUUUUGUGCUAAAUUGUUUUGGCACGCAAAUAAAUGGCAGGA --((((..((.(((...------...))).))..)))).....((((....((((((.......))))))..(((((.(((((((((....))))))))).....))))))))).. ( -31.10) >DroPer_CAF1 55467 99 - 1 --GUGUUUG------UC------CG---UGUGUCAAAGGAUAACUGCAGUCGUGCAUAUUUUAAAUGCGCACAAUUUUUGUGUUAAAUUGUUUUGGCACGCAAAUAAAUGGCAGGA --....(((------((------(.---.........))))))((((.((.((((((.......)))))))).((((.(((((((((....))))))))).)))).....)))).. ( -27.60) >consensus __GUGUUUG______UC______CG___CGUGUCAAAGGAUAACUGCAGUCGUGCAUAUUUUAAAUGCGCACAAUUUUUGUGUUAAAUUGUUUUGGCACGCAAAUAAAUGGCAGGA ..............................((((....)))).((((....((((((.......))))))...((((.(((((((((....))))))))).)))).....)))).. (-18.47 = -19.25 + 0.78)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:26 2006