
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,730,151 – 26,730,265 |
| Length | 114 |
| Max. P | 0.867629 |

| Location | 26,730,151 – 26,730,265 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.50 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -20.69 |
| Energy contribution | -20.08 |
| Covariance contribution | -0.61 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.867629 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26730151 114 - 27905053 GCCU---CUUCCGGUUUGUUUGUUGCGCGCAUUAAGCUAAUUAUGGCCAAAUAAAGUGAGACGAAACGAAAAGCGCGCUUCGAGCAAACAGAGGAAUAUAUC-C--GAAUCCUCUAUCCC ....---......(((((((((..((((((.....((((....))))........((........)).....))))))..)))))))))((((((.......-.--...))))))..... ( -33.90) >DroPse_CAF1 42720 113 - 1 UGCU---CUUCCUGGUUGUUUGUUGUGCGCAUUAAGCUAAUUAUGGCCAAAUAAAGUGAGACGAAAGGAGACGCGCGCUCUGGGCAAAUAGCAGGGUGGAGA-A---GGUCCUGUAUCCU ..((---((.(((.((((((((((((((((.....((((....))))...................(....)))))))....)))))))))).))).)))).-(---((........))) ( -34.90) >DroWil_CAF1 38744 113 - 1 UCCU---CUUUCUGGUUGUUUGUUCUGCGCAUUAAGCUAAUUAUGGCCAAAUAAAGUGAAACGAAAUGAAACGCGUGCACCGAACAAACAGCAACAUGGAAC-AACAAUUUCUCU---CU (((.---.......((((((((((((((((.....((((....))))........(((.............))))))))..))))))))))).....)))..-............---.. ( -26.14) >DroMoj_CAF1 35446 108 - 1 UGCUAGAUAUCCUGGUUGUUUGCUUCGCGCAUUAAGCUAAUUAUGGCCAAAUAAAGCGGCUCGAAUCGAAGCGCGCGCUCUGUGCAAAUAGCAGA-UGAAAC-A--GAG----CUA---- .(((...((((...(((((((((...((((.....((((....))))........(((..(((...)))..))))))).....))))))))).))-))....-.--.))----)..---- ( -29.60) >DroAna_CAF1 41429 111 - 1 GGC----CGUUCGUUUUGUUUGUUGCGCGCAUUAAGCUAAUUAUGGCCAAAUAAAGCGAGACGAAUUGAAACGCGCGCUCUAAGCAAACAG---GAUAUAUUCA--GUAUCCUCUGUUGC (..----(((.((((((((((((..(((..(((..((((....))))..)))...)))..))))).))))))).)))..)...((((..((---(((((.....--)))))))...)))) ( -32.30) >DroPer_CAF1 40868 113 - 1 UGCU---CUUCCUGGUUGUUUGUUGUGCGCAUUAAGCUAAUUAUGGCCAAAUAAAGUGAGACGAAAGGAGACGCGCGCUCUGGGCAAACAGCAGGGUGGAGA-A---GGUCCUGUAUCCU ..((---((.(((.((((((((((((((((.....((((....))))...................(....)))))))....)))))))))).))).)))).-(---((........))) ( -36.80) >consensus UGCU___CUUCCUGGUUGUUUGUUGCGCGCAUUAAGCUAAUUAUGGCCAAAUAAAGUGAGACGAAACGAAACGCGCGCUCUGAGCAAACAGCAGGAUGGAAC_A__GAGUCCUCUAUCCU ..............((((((((((((((((.....((((....))))........(.....)..........))))))....))))))))))............................ (-20.69 = -20.08 + -0.61)

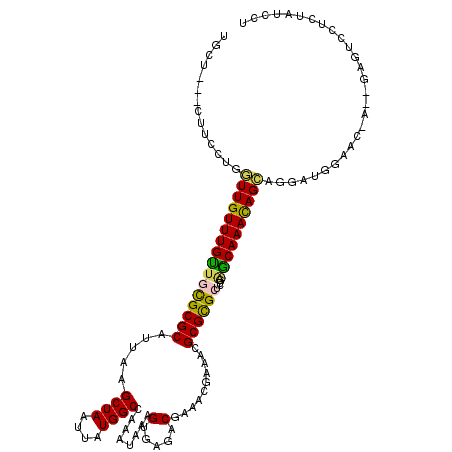
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:21 2006