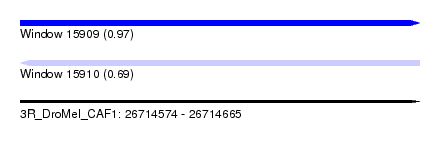
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,714,574 – 26,714,665 |
| Length | 91 |
| Max. P | 0.971211 |

| Location | 26,714,574 – 26,714,665 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 73.55 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -16.58 |
| Energy contribution | -16.97 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.971211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26714574 91 + 27905053 ------UUCGCACUUC---UCUCCAUCU---CUUCUCCACUCACCAAAGGCGCAGCACAGUGCCAAAACGGCCAAGAGGCCUGCUCUCUCCAUUUUCCCGGUU ------..........---.........---...........(((...(((((......))))).....((((....))))..................))). ( -18.80) >DroPse_CAF1 22914 87 + 1 ACGCAAUUUGCACCUA---U-------G---ACACACAACUUACCGCAGGCACAA---ACAGCCAAAAUGGCCAAAAGGCCAGCUCUGCGCAUUUUGUUGAUU ..((.....)).....---.-------.---.....((((....(((((((....---...)))....(((((....)))))....))))......))))... ( -22.10) >DroSec_CAF1 18181 83 + 1 ------UUCGCACUUC---U-----------CUUCUGCACUCACCGAAGGCGCAGCACAGUGCCAAAACGGCCAAGAGGCCUGCUCUCUCCAUUUUCCCGGUU ------...(((....---.-----------....)))....((((..(((((......))))).....((((....)))).................)))). ( -21.90) >DroSim_CAF1 18379 83 + 1 ------UUCGCACUUU---U-----------CUUCUGCACUCACCGAAGGCGCAGCACAGUGCCAAAACGGCCAAGAGGCCUGCUCUCUCCAUUUUCCCGGUU ------...(((....---.-----------....)))....((((..(((((......))))).....((((....)))).................)))). ( -21.90) >DroEre_CAF1 17802 76 + 1 ------UUC---------------------GCUAUCGCACUCACCGAAGGCGCAGCACAGUGCCAAAACGGCCAAAAGGCCUGCUCUCUCCAUGUUCCCGGUU ------...---------------------((....))....((((..(((((......))))).....((((....)))).................)))). ( -21.80) >DroYak_CAF1 18029 97 + 1 ------UUCGUAUUUCGUCUUUCGCUCUUUGCUUUCGCACUCACCGAAGGCACAGCACAGUGCCAAAACGGCCAAAAGGCCUGCUCUCUCCAUGUUCCGGGUU ------.................((((..((((((((.......)))))))).((((..((......))((((....)))))))).............)))). ( -22.30) >consensus ______UUCGCACUUC___U___________CUUCUGCACUCACCGAAGGCGCAGCACAGUGCCAAAACGGCCAAAAGGCCUGCUCUCUCCAUUUUCCCGGUU ..........................................(((...(((((......))))).....((((....))))..................))). (-16.58 = -16.97 + 0.39)

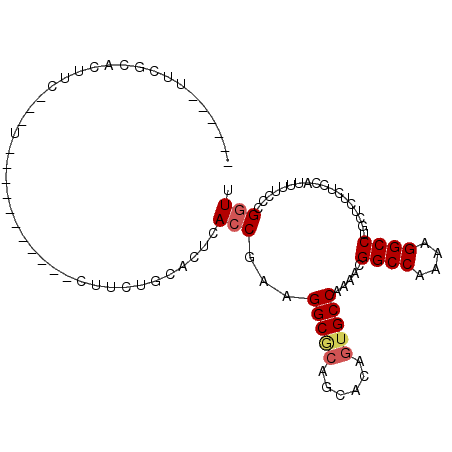
| Location | 26,714,574 – 26,714,665 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 73.55 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -17.07 |
| Energy contribution | -17.85 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.686299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26714574 91 - 27905053 AACCGGGAAAAUGGAGAGAGCAGGCCUCUUGGCCGUUUUGGCACUGUGCUGCGCCUUUGGUGAGUGGAGAAG---AGAUGGAGA---GAAGUGCGAA------ ..(((......)))..(((((.((((....))))))))).(((((...((.((.((((............))---)).)).)).---..)))))...------ ( -25.50) >DroPse_CAF1 22914 87 - 1 AAUCAACAAAAUGCGCAGAGCUGGCCUUUUGGCCAUUUUGGCUGU---UUGUGCCUGCGGUAAGUUGUGUGU---C-------A---UAGGUGCAAAUUGCGU ......((((((((.....)).((((....))))))))))((.((---(((..((((.(..(.......)..---)-------.---))))..))))).)).. ( -29.00) >DroSec_CAF1 18181 83 - 1 AACCGGGAAAAUGGAGAGAGCAGGCCUCUUGGCCGUUUUGGCACUGUGCUGCGCCUUCGGUGAGUGCAGAAG-----------A---GAAGUGCGAA------ ..(((......)))..(((((.((((....))))))))).(((((...(((((((......).))))))...-----------.---..)))))...------ ( -29.10) >DroSim_CAF1 18379 83 - 1 AACCGGGAAAAUGGAGAGAGCAGGCCUCUUGGCCGUUUUGGCACUGUGCUGCGCCUUCGGUGAGUGCAGAAG-----------A---AAAGUGCGAA------ ..(((......)))..(((((.((((....))))))))).(((((...(((((((......).))))))...-----------.---..)))))...------ ( -29.10) >DroEre_CAF1 17802 76 - 1 AACCGGGAACAUGGAGAGAGCAGGCCUUUUGGCCGUUUUGGCACUGUGCUGCGCCUUCGGUGAGUGCGAUAGC---------------------GAA------ .((((((.........(((((.((((....)))))))))(((.(......).)))))))))...(((....))---------------------)..------ ( -23.60) >DroYak_CAF1 18029 97 - 1 AACCCGGAACAUGGAGAGAGCAGGCCUUUUGGCCGUUUUGGCACUGUGCUGUGCCUUCGGUGAGUGCGAAAGCAAAGAGCGAAAGACGAAAUACGAA------ ....((...(((.((((((((.((((....)))))))))(((((......)))))))).)))..(((....)))....((....).)......))..------ ( -31.60) >consensus AACCGGGAAAAUGGAGAGAGCAGGCCUCUUGGCCGUUUUGGCACUGUGCUGCGCCUUCGGUGAGUGCAGAAG___________A___GAAGUGCGAA______ .((((((.........(((((.((((....)))))))))(((.(......).))))))))).......................................... (-17.07 = -17.85 + 0.78)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:18 2006