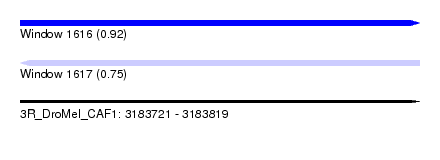
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,183,721 – 3,183,819 |
| Length | 98 |
| Max. P | 0.919644 |

| Location | 3,183,721 – 3,183,819 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -23.26 |
| Energy contribution | -22.87 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919644 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3183721 98 + 27905053 ---GCCGGAAAACCUUGGAAAAAAG-GCAGCAAAGCGUUAAGCCGUUAGCAGCUAGAUUGAGCCCAGCAGCUGCUGGCGACACCUUGCAACAGUUGCCU--G---------------AA- ---.((((......)))).....((-(((((...((.....))(((((((((((.(.(((....))))))))))))))).............)))))))--.---------------..- ( -33.00) >DroVir_CAF1 4275 117 + 1 AACAUG-GAUAGCACAAGAAAAAAGCGCAGCACAGCGUCAAACCGUUGGCGGUUGCGUUAAGC--AACAGCUGCCAGCGACACCUUGCAAGAGUUGCUGUUGUAAAGUGUUGGGCGGAAA ....((-..((((((........((((((((.(((((......)))))...))))))))..((--((((((.(((.((((....))))..).)).))))))))...))))))..)).... ( -39.50) >DroSec_CAF1 3065 98 + 1 ---GCCGGAAAACCUUGGAAAAAAG-GCAGCAAAGCGUUAAACCGUUAGCAGCUAGAUUGAGCCCAGCAGCUGCUGGCGACACCUUGCAACAGUUGCCU--G---------------AA- ---.((((......)))).....((-(((((...(((......(((((((((((.(.(((....)))))))))))))))......)))....)))))))--.---------------..- ( -32.70) >DroSim_CAF1 9097 98 + 1 ---GCCGGAAAACCUUGGAAAAAAG-GCAGCAAAGCGUUAAACCGUUAGCAGCUAGAUUGAGCCCAGCAGCUGCUGGCGACACCUUGCAACAGUUGCCU--G---------------AA- ---.((((......)))).....((-(((((...(((......(((((((((((.(.(((....)))))))))))))))......)))....)))))))--.---------------..- ( -32.70) >DroEre_CAF1 3932 95 + 1 ---GCCGGAAGUCCUUGGAAAAAAG-GCAGCAAAGCAUUAAACCGUUA---GCUAGAUUGAGCCCAGCAGCUGCUGGCGACACCUUGCAACAGUUGCCU--G---------------AG- ---.((((......)))).....((-(((((...(((......(((((---((.((.(((.......))))))))))))......)))....)))))))--.---------------..- ( -25.30) >DroYak_CAF1 8904 104 + 1 ---GCCGGGAAUCCUUGGAA-AAAG-GCAGCAAAGUGUUAAACCGUUAGCAGCUAGAUUGAGCCGAGCAGCUGCUGGCGACACCUUGCAACAGUUGCCU--GGAAAA--------GGAA- ---.(((((...((((....-.)))-)..((((.((((.....(((((((((((...(((...)))..))))))))))))))).))))........)))--))....--------....- ( -35.90) >consensus ___GCCGGAAAACCUUGGAAAAAAG_GCAGCAAAGCGUUAAACCGUUAGCAGCUAGAUUGAGCCCAGCAGCUGCUGGCGACACCUUGCAACAGUUGCCU__G_______________AA_ ....((((......))))........(((((...(((......(((((((((((..............)))))))))))......)))....)))))....................... (-23.26 = -22.87 + -0.38)


| Location | 3,183,721 – 3,183,819 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -31.60 |
| Consensus MFE | -21.45 |
| Energy contribution | -21.54 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745164 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3183721 98 - 27905053 -UU---------------C--AGGCAACUGUUGCAAGGUGUCGCCAGCAGCUGCUGGGCUCAAUCUAGCUGCUAACGGCUUAACGCUUUGCUGC-CUUUUUUCCAAGGUUUUCCGGC--- -..---------------(--((....)))..(((((((((.(((...(((.((((((.....)))))).)))...)))...))))))))).((-(((......)))))........--- ( -34.20) >DroVir_CAF1 4275 117 - 1 UUUCCGCCCAACACUUUACAACAGCAACUCUUGCAAGGUGUCGCUGGCAGCUGUU--GCUUAACGCAACCGCCAACGGUUUGACGCUGUGCUGCGCUUUUUUCUUGUGCUAUC-CAUGUU ......................((((......(((.((((((((((...((.(((--((.....))))).))...))))..)))))).))).((((.........))))....-..)))) ( -29.90) >DroSec_CAF1 3065 98 - 1 -UU---------------C--AGGCAACUGUUGCAAGGUGUCGCCAGCAGCUGCUGGGCUCAAUCUAGCUGCUAACGGUUUAACGCUUUGCUGC-CUUUUUUCCAAGGUUUUCCGGC--- -..---------------(--((....)))..(((((((((.(((...(((.((((((.....)))))).)))...)))...))))))))).((-(((......)))))........--- ( -32.60) >DroSim_CAF1 9097 98 - 1 -UU---------------C--AGGCAACUGUUGCAAGGUGUCGCCAGCAGCUGCUGGGCUCAAUCUAGCUGCUAACGGUUUAACGCUUUGCUGC-CUUUUUUCCAAGGUUUUCCGGC--- -..---------------(--((....)))..(((((((((.(((...(((.((((((.....)))))).)))...)))...))))))))).((-(((......)))))........--- ( -32.60) >DroEre_CAF1 3932 95 - 1 -CU---------------C--AGGCAACUGUUGCAAGGUGUCGCCAGCAGCUGCUGGGCUCAAUCUAGC---UAACGGUUUAAUGCUUUGCUGC-CUUUUUUCCAAGGACUUCCGGC--- -..---------------.--(((((((((((((.((((...((((((....))).)))...)))).))---.))))))....))))).(((((-(((......)))).....))))--- ( -26.90) >DroYak_CAF1 8904 104 - 1 -UUCC--------UUUUCC--AGGCAACUGUUGCAAGGUGUCGCCAGCAGCUGCUCGGCUCAAUCUAGCUGCUAACGGUUUAACACUUUGCUGC-CUUU-UUCCAAGGAUUCCCGGC--- -.(((--------((....--(((((......(((((((((.(((((((((((............))))))))...)))...))))))))))))-))..-....)))))........--- ( -33.40) >consensus _UU_______________C__AGGCAACUGUUGCAAGGUGUCGCCAGCAGCUGCUGGGCUCAAUCUAGCUGCUAACGGUUUAACGCUUUGCUGC_CUUUUUUCCAAGGUUUUCCGGC___ ......................((.....((.(((((((((.(((...(((.((((((.....)))))).)))...)))...))))))))).))........))................ (-21.45 = -21.54 + 0.09)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:59 2006