
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,695,897 – 26,696,013 |
| Length | 116 |
| Max. P | 0.742476 |

| Location | 26,695,897 – 26,695,987 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 81.30 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -17.96 |
| Energy contribution | -18.85 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742476 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26695897 90 - 27905053 UUCGCAA-AUCCGAACGAAAAUCCGCAAUUGAGCAAGCCAAUUCAA------------UCGAUUGCGAUUGCCGCAAAAGUGGGUGCACAAAGGCACGCGGAG----- (((((..-...............(((((((((..............------------)))))))))....((((....))))((((......))))))))).----- ( -26.54) >DroSec_CAF1 2296 90 - 1 UUCGCAA-AUCCGAACGAAAAUCCGCAAUUGAGCAAGCCAAUUCCA------------UCGAUUGCGAUUGCCGCAAAAGUGGGUGCACAAAGGCACGCGGAG----- (((((..-...............(((((((((..............------------)))))))))....((((....))))((((......))))))))).----- ( -26.54) >DroEre_CAF1 2304 90 - 1 UUCGCAA-AUCCGAACGAAAAUCCGCAAUUGAGCAAGCCAAUUCAA------------UCGAUUGCGAUUGCCGCAAAAGUGGGUGCACAAAGGCACGCGGAG----- (((((..-...............(((((((((..............------------)))))))))....((((....))))((((......))))))))).----- ( -26.54) >DroYak_CAF1 2317 90 - 1 UUCGCAA-AUGCGAACGAAAAUCCGCAAUUGAGCAAGCAAAUUCAA------------UCGAUUGCGAUUGCCGCAAAAGUGGGUGCAGAAAGGCAAGCGGAG----- (((((..-..)))))......(((((.................(((------------(((....))))))((((....)))).(((......))).))))).----- ( -26.00) >DroAna_CAF1 18889 90 - 1 UUCGCAAAACCCGAACUAAAAUCCGCAAUUGGCUUGCUCAAACCAA------------UCCAAUUCGAUUGCCGCAA-AGUGGGUGCAAAAAGGCACGUGAAG----- (((((..................((.((((((.(((.......)))------------.))))))))....((((..-.))))((((......))))))))).----- ( -20.30) >DroPer_CAF1 18451 108 - 1 UUCGCAAAAGCCGAACGAAAAUCCGCCAUUGAGUAAGCGAAUCCGAGUCGUAGUCCACGCGAUUGCGAUUGCCACAAAAGUGGGUGCAAAAAAGUACUAGCAAAACAA (((((....(((((.((......))...))).))..)))))....((((((((((.....)))))))))).((((....))))((((......))))........... ( -23.90) >consensus UUCGCAA_AUCCGAACGAAAAUCCGCAAUUGAGCAAGCCAAUUCAA____________UCGAUUGCGAUUGCCGCAAAAGUGGGUGCACAAAGGCACGCGGAG_____ (((((..................(((((((((..........................)))))))))....((((....))))((((......)))))))))...... (-17.96 = -18.85 + 0.89)

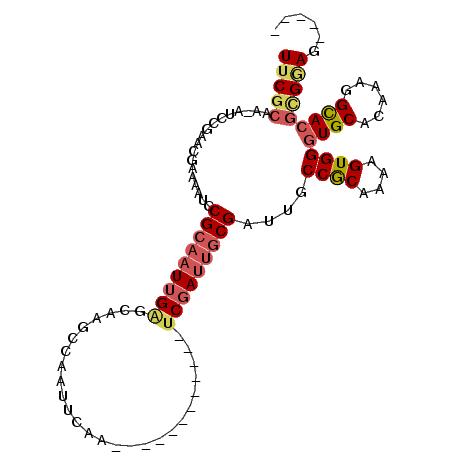
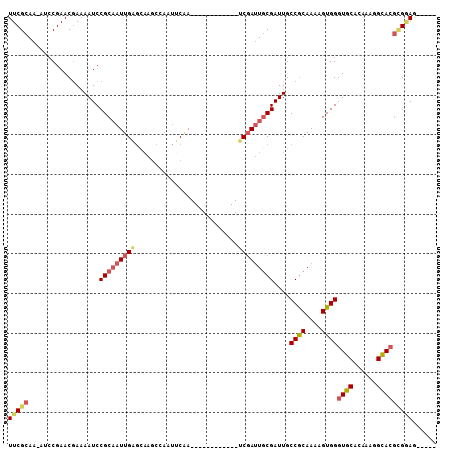
| Location | 26,695,923 – 26,696,013 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.34 |
| Mean single sequence MFE | -20.73 |
| Consensus MFE | -13.00 |
| Energy contribution | -14.09 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.63 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.513604 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26695923 90 - 27905053 ----CAGGCAAAUUCUCGACUCUUACCUCUUUCGCAA-AUCCGAACGAAAAUCCGCAAUUGAGCAAGCCAAUUCAA------------UCGAUUGCGAUUGCCGCAA ----..(((((................((.((((...-...)))).)).....(((((((((..............------------))))))))).))))).... ( -20.94) >DroSec_CAF1 2322 90 - 1 ----CAGGCAAAUUCUCGACUCUUACCUCUUUCGCAA-AUCCGAACGAAAAUCCGCAAUUGAGCAAGCCAAUUCCA------------UCGAUUGCGAUUGCCGCAA ----..(((((................((.((((...-...)))).)).....(((((((((..............------------))))))))).))))).... ( -20.94) >DroEre_CAF1 2330 90 - 1 ----CAGGCAAAUUCUCGACUCUUACCUCUUUCGCAA-AUCCGAACGAAAAUCCGCAAUUGAGCAAGCCAAUUCAA------------UCGAUUGCGAUUGCCGCAA ----..(((((................((.((((...-...)))).)).....(((((((((..............------------))))))))).))))).... ( -20.94) >DroYak_CAF1 2343 90 - 1 ----CAGGUAAAUUCUCGACUUUUACCUCUUUCGCAA-AUGCGAACGAAAAUCCGCAAUUGAGCAAGCAAAUUCAA------------UCGAUUGCGAUUGCCGCAA ----..(((((................((.(((((..-..))))).)).....(((((((((..............------------))))))))).))))).... ( -23.24) >DroAna_CAF1 18914 95 - 1 GUGUCAGGCAAAUUCUUGACUUUUACCUCUUUCGCAAAACCCGAACUAAAAUCCGCAAUUGGCUUGCUCAAACCAA------------UCCAAUUCGAUUGCCGCAA ......(((((...................((((.......))))........((.((((((.(((.......)))------------.)))))))).))))).... ( -15.70) >DroPer_CAF1 18482 107 - 1 AUGGCCAGCACAUUCUUGACUUUUACCUCUUUCGCAAAAGCCGAACGAAAAUCCGCCAUUGAGUAAGCGAAUCCGAGUCGUAGUCCACGCGAUUGCGAUUGCCACAA .((((..((..((((............((.(((((....).)))).))............))))..)).......((((((((((.....))))))))))))))... ( -22.65) >consensus ____CAGGCAAAUUCUCGACUCUUACCUCUUUCGCAA_AUCCGAACGAAAAUCCGCAAUUGAGCAAGCCAAUUCAA____________UCGAUUGCGAUUGCCGCAA ......(((((................((.((((.......)))).)).....(((((((((..........................))))))))).))))).... (-13.00 = -14.09 + 1.08)

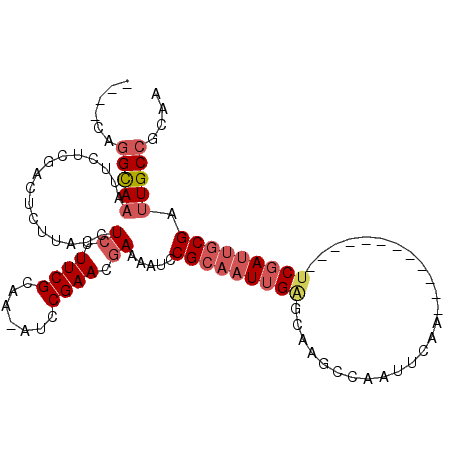
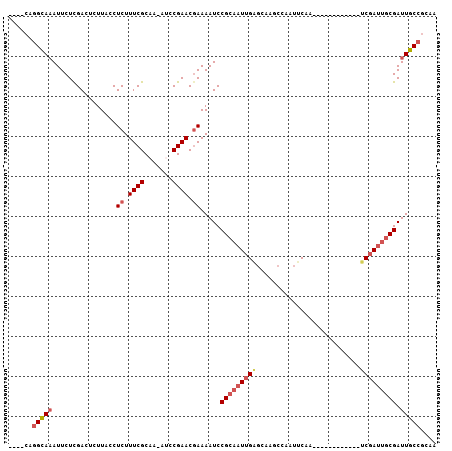
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:14 2006