
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,692,132 – 26,692,270 |
| Length | 138 |
| Max. P | 0.980730 |

| Location | 26,692,132 – 26,692,230 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -40.30 |
| Consensus MFE | -30.02 |
| Energy contribution | -30.55 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.698591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26692132 98 + 27905053 ACAUUACGCAAAGUGGGGG--------GCGUGGGAGUGGGCGUGUG-------GC-------AUGUGGGUGGGCGGCCAACUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUU ...((((((..........--------))))))........((.((-------((-------((.((..(((((((((((.....))))))........)))))..)))))))).))... ( -36.90) >DroSec_CAF1 282827 104 + 1 ACAUUACGCAAAGUGG-GG--------GCGUGGGAGUGGGCGUGUG-------GCAUGUGGCAUGUGGGUGGGCGGCCAACUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUU ...........((((.-.(--------((((.(...(((((.((((-------((....((((.((.(((.....))).))))))...)))))).....)))))..).)))))...)))) ( -38.10) >DroSim_CAF1 279596 105 + 1 ACAUUACGCAAAGUGGGGG--------GCGUGGGAGUGGGCGUGUG-------GCAUGUGGCAUGUGGGUGGGCGGCCAACUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUU ...........((((...(--------((((.(...(((((.((((-------((....((((.((.(((.....))).))))))...)))))).....)))))..).)))))...)))) ( -38.40) >DroEre_CAF1 298860 120 + 1 ACAUUACGCAAAGUGUGGGGCGGUGAGGCGUGGGAGUGGGCGUGUGGCAUGUGGCAUGUGGCAUGUGGGUGGGCGGCCAAGUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUU .(((..(.(........).)..))).(((((.(...(((((.((((((....(((((.((((.(((......))))))).)))))...)))))).....)))))..).)))))....... ( -45.40) >DroYak_CAF1 287960 118 + 1 ACAUUACGCAAAGUGUGGGGCGGUGAGGCGUGGGAGUGGGCGU--GGAAGUGGGCGUGUGGCAUGUGGGUGGGCGGCCAAGUGCUGUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUU ...((((((..(.(.(.(.((......)).).).).)..))))--))........((.((((((.((..((((((((((.......)))))........)))))..)))))))).))... ( -42.70) >consensus ACAUUACGCAAAGUGGGGG________GCGUGGGAGUGGGCGUGUG_______GCAUGUGGCAUGUGGGUGGGCGGCCAACUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUU ...((((((..................))))))..((.(((((..........(((((...)))))(..(((((((((((.....))))))........)))))..).)))))..))... (-30.02 = -30.55 + 0.53)


| Location | 26,692,132 – 26,692,230 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -30.10 |
| Consensus MFE | -23.99 |
| Energy contribution | -24.99 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.725090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26692132 98 - 27905053 AAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCAGUUGGCCGCCCACCCACAU-------GC-------CACACGCCCACUCCCACGC--------CCCCCACUUUGCGUAAUGU ...((.((((((((..(((((........((((((.....)))))))))))..)).))-------))-------)).))..........((((--------..........))))..... ( -30.70) >DroSec_CAF1 282827 104 - 1 AAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCAGUUGGCCGCCCACCCACAUGCCACAUGC-------CACACGCCCACUCCCACGC--------CC-CCACUUUGCGUAAUGU .((((..(((((((..(((((........((((((.....)))))))))))..)).)))))))))..-------...............((((--------..-.......))))..... ( -29.10) >DroSim_CAF1 279596 105 - 1 AAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCAGUUGGCCGCCCACCCACAUGCCACAUGC-------CACACGCCCACUCCCACGC--------CCCCCACUUUGCGUAAUGU .((((..(((((((..(((((........((((((.....)))))))))))..)).)))))))))..-------...............((((--------..........))))..... ( -29.00) >DroEre_CAF1 298860 120 - 1 AAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCACUUGGCCGCCCACCCACAUGCCACAUGCCACAUGCCACACGCCCACUCCCACGCCUCACCGCCCCACACUUUGCGUAAUGU .((((..(((((((..(((((........((((((.....)))))))))))..)).)))))))))...((((.....((((..........((......))..........)))).)))) ( -30.95) >DroYak_CAF1 287960 118 - 1 AAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCACAGCACUUGGCCGCCCACCCACAUGCCACACGCCCACUUCC--ACGCCCACUCCCACGCCUCACCGCCCCACACUUUGCGUAAUGU ...((.((((((((..(((((........(((((.......))))))))))..)).)))))).))..........--((((..........((......))..........))))..... ( -30.75) >consensus AAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCAGUUGGCCGCCCACCCACAUGCCACAUGC_______CACACGCCCACUCCCACGC________CCCCCACUUUGCGUAAUGU ......((((((((..(((((........((((((.....)))))))))))..)).))))))...........................((((..................))))..... (-23.99 = -24.99 + 1.00)

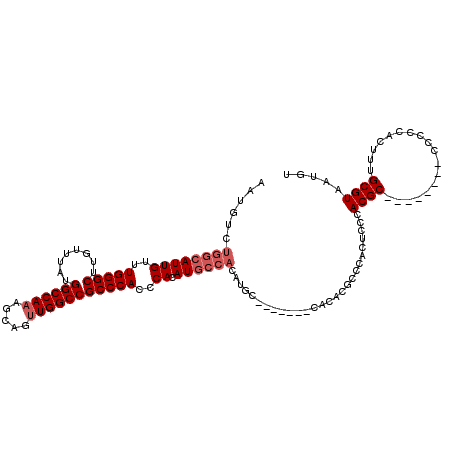
| Location | 26,692,164 – 26,692,270 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -29.12 |
| Energy contribution | -30.12 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980730 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26692164 106 + 27905053 CGUGUG-------GC-------AUGUGGGUGGGCGGCCAACUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUUUCAAACACAAUAAAUAAUUGAAGCCAGCGCAGAGUCAACA .((.((-------((-------((.((..(((((((((((.....))))))........)))))..)))))))).))..(((.....((((.....))))..((....)).)))...... ( -33.70) >DroSec_CAF1 282858 113 + 1 CGUGUG-------GCAUGUGGCAUGUGGGUGGGCGGCCAACUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUUUCAAACACAAUAAAUAAUUGAAGCCAGCGCAGAGUCAACA .(((((-------((.(.((((((.((..(((((((((((.....))))))........)))))..)))))))).)...........((((.....))))..)))).))).......... ( -39.00) >DroSim_CAF1 279628 113 + 1 CGUGUG-------GCAUGUGGCAUGUGGGUGGGCGGCCAACUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUUUCAAACACAAUAAAUAAUUGAAGCCAGCGCAGAGUCAACA .(((((-------((.(.((((((.((..(((((((((((.....))))))........)))))..)))))))).)...........((((.....))))..)))).))).......... ( -39.00) >DroEre_CAF1 298900 120 + 1 CGUGUGGCAUGUGGCAUGUGGCAUGUGGGUGGGCGGCCAAGUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUUUCAAACACAAUAAAUAAUUGAAGCCAGCGCAGAGUCAACA .(((((((...(((.(((((((((.((..((((((((((((...)))))))........)))))..)))))))..)))).)))....((((.....))))..)))).))).......... ( -40.50) >DroYak_CAF1 288000 118 + 1 CGU--GGAAGUGGGCGUGUGGCAUGUGGGUGGGCGGCCAAGUGCUGUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUUUCAAACACAAUAAAUAAUUGAAGCCAGCGCAGAGUCAACA .((--.((.((((((((.((((((.((..((((((((((.......)))))........)))))..)))))))).))..........((((.....))))..)))..)))....)).)). ( -39.10) >consensus CGUGUG_______GCAUGUGGCAUGUGGGUGGGCGGCCAACUGCUUUGGCCAUAAACAAGCCCAAACAAUGCCAGACAUUUCAAACACAAUAAAUAAUUGAAGCCAGCGCAGAGUCAACA ..................((((((.((..(((((((((((.....))))))........)))))..))))))))(((..........((((.....))))..((....))...))).... (-29.12 = -30.12 + 1.00)



| Location | 26,692,164 – 26,692,270 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -34.14 |
| Consensus MFE | -27.90 |
| Energy contribution | -28.90 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736328 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26692164 106 - 27905053 UGUUGACUCUGCGCUGGCUUCAAUUAUUUAUUGUGUUUGAAAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCAGUUGGCCGCCCACCCACAU-------GC-------CACACG ....(((...((....))(((((.(((.....))).)))))..)))((((((((..(((((........((((((.....)))))))))))..)).))-------))-------)).... ( -31.40) >DroSec_CAF1 282858 113 - 1 UGUUGACUCUGCGCUGGCUUCAAUUAUUUAUUGUGUUUGAAAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCAGUUGGCCGCCCACCCACAUGCCACAUGC-------CACACG ..........(.(.(((((((((.(((.....))).)))))((((..(((((((..(((((........((((((.....)))))))))))..)).)))))))))))-------))).). ( -34.70) >DroSim_CAF1 279628 113 - 1 UGUUGACUCUGCGCUGGCUUCAAUUAUUUAUUGUGUUUGAAAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCAGUUGGCCGCCCACCCACAUGCCACAUGC-------CACACG ..........(.(.(((((((((.(((.....))).)))))((((..(((((((..(((((........((((((.....)))))))))))..)).)))))))))))-------))).). ( -34.70) >DroEre_CAF1 298900 120 - 1 UGUUGACUCUGCGCUGGCUUCAAUUAUUUAUUGUGUUUGAAAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCACUUGGCCGCCCACCCACAUGCCACAUGCCACAUGCCACACG ..........(((.(((((((((.(((.....))).)))))((((..(((((((..(((((........((((((.....)))))))))))..)).)))))))))))))..)))...... ( -36.30) >DroYak_CAF1 288000 118 - 1 UGUUGACUCUGCGCUGGCUUCAAUUAUUUAUUGUGUUUGAAAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCACAGCACUUGGCCGCCCACCCACAUGCCACACGCCCACUUCC--ACG ...............((((((((.(((.....))).)))))..((.((((((((..(((((........(((((.......))))))))))..)).)))))).))))).......--... ( -33.60) >consensus UGUUGACUCUGCGCUGGCUUCAAUUAUUUAUUGUGUUUGAAAUGUCUGGCAUUGUUUGGGCUUGUUUAUGGCCAAAGCAGUUGGCCGCCCACCCACAUGCCACAUGC_______CACACG ....(((...((....))(((((.(((.....))).)))))..)))((((((((..(((((........((((((.....)))))))))))..)).)))))).................. (-27.90 = -28.90 + 1.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:12 2006