
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,690,535 – 26,690,651 |
| Length | 116 |
| Max. P | 0.692885 |

| Location | 26,690,535 – 26,690,651 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -15.60 |
| Energy contribution | -16.20 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692885 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26690535 116 + 27905053 AUCUCCCCA---UCUCAAACUCCUGGCCUUCUCCUUGCUCGCUCUCUUUUCCACCAGGAAUAAAUGGAUCGAAUUUCCCGACGAGUUUGGCGAGUU-UGGUUUUGACUUCUGAUGGAAAU ......(((---(((((((..((..((..((.((..(((((.((....((((....)))).....(((.......))).)))))))..)).)))).-.)).))))).....))))).... ( -29.30) >DroSec_CAF1 281261 103 + 1 AUCUCCCCAUCAUCUCAAACUACUUGCCU----------------CCUUUCCACCAGGAAUAAAUGGAUCGUAUUUCCCGACGAGUUUGGCGAGUU-UGGUUUUGACUUCUGAUGGAAAU ......((((((..(((((((((((((((----------------((.((((....)))).....)))((((........))))....))))))).-.)).)))))....)))))).... ( -29.00) >DroSim_CAF1 278026 103 + 1 AUCUCCCCAUCAUCUGAAACUCCUUGCCU----------------CCUUUCCACCAGGAAUAAAUGGAUCGUAUUUCCCGACGAGUUUGGCGAGCU-UGGUUUUGACUUCUGAUGGAAAU ......((((((((.((((((.(((((((----------------((.((((....)))).....)))((((........))))....))))))..-.))))))))....)))))).... ( -27.60) >DroEre_CAF1 297279 113 + 1 AUCUCCCCAUC-UCUCAAAGUCCUUAUU------UCGCUCGCUCUCUUUUCCACCAGGAAUAAAUGGAUCGUAUUUCCCGACGAGUUUAGCGAGUUUUGGUUUUGACUUCUGAUGGAAAU ......(((((-..((((((.((....(------(((((.((((((..((((....)))).....(((.......))).)).))))..))))))....)))))))).....))))).... ( -30.20) >DroYak_CAF1 286315 111 + 1 AUCUCCCCAUC---UCAAAGUCCUUAUU------UCGCUCGCUCUCUUUUCCACCAGGAAUAAAUGGAUCGUAUUUCCCGACGAGUUUAGCGAGUUUUGGCUUUGACUUCUGAUGGAAAU ......(((((---((((((.((....(------(((((.((((((..((((....)))).....(((.......))).)).))))..))))))....)))))))).....))))).... ( -31.80) >consensus AUCUCCCCAUC_UCUCAAACUCCUUGCCU_____U_GCUCGCUCUCUUUUCCACCAGGAAUAAAUGGAUCGUAUUUCCCGACGAGUUUGGCGAGUU_UGGUUUUGACUUCUGAUGGAAAU ...............................................((((((.(((((......((((((...(((((((.....)))).)))...))))))....))))).)))))). (-15.60 = -16.20 + 0.60)


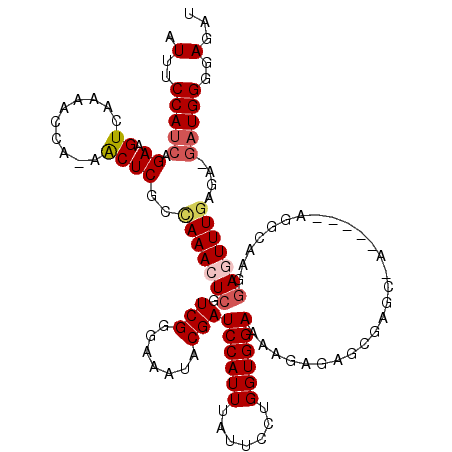
| Location | 26,690,535 – 26,690,651 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.20 |
| Mean single sequence MFE | -30.50 |
| Consensus MFE | -18.02 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.505827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26690535 116 - 27905053 AUUUCCAUCAGAAGUCAAAACCA-AACUCGCCAAACUCGUCGGGAAAUUCGAUCCAUUUAUUCCUGGUGGAAAAGAGAGCGAGCAAGGAGAAGGCCAGGAGUUUGAGA---UGGGGAGAU .(..(((((.(....).....((-(((((......((((((.(((.......))).(((.((((....)))).))))).))))...((......))..))))))).))---)))..)... ( -30.40) >DroSec_CAF1 281261 103 - 1 AUUUCCAUCAGAAGUCAAAACCA-AACUCGCCAAACUCGUCGGGAAAUACGAUCCAUUUAUUCCUGGUGGAAAGG----------------AGGCAAGUAGUUUGAGAUGAUGGGGAGAU .(..((((((....(((((....-.(((.(((.......(((.......)))((((((.......))))))....----------------.))).)))..)))))..))))))..)... ( -28.10) >DroSim_CAF1 278026 103 - 1 AUUUCCAUCAGAAGUCAAAACCA-AGCUCGCCAAACUCGUCGGGAAAUACGAUCCAUUUAUUCCUGGUGGAAAGG----------------AGGCAAGGAGUUUCAGAUGAUGGGGAGAU .(..(((((((....)......(-((((((((.......(((.......)))((((((.......))))))....----------------.)))...))))))....))))))..)... ( -25.50) >DroEre_CAF1 297279 113 - 1 AUUUCCAUCAGAAGUCAAAACCAAAACUCGCUAAACUCGUCGGGAAAUACGAUCCAUUUAUUCCUGGUGGAAAAGAGAGCGAGCGA------AAUAAGGACUUUGAGA-GAUGGGGAGAU .(..(((((.((((((..........((((((...(((.(((.......)))((((((.......))))))...)))))))))...------......))))))....-)))))..)... ( -33.71) >DroYak_CAF1 286315 111 - 1 AUUUCCAUCAGAAGUCAAAGCCAAAACUCGCUAAACUCGUCGGGAAAUACGAUCCAUUUAUUCCUGGUGGAAAAGAGAGCGAGCGA------AAUAAGGACUUUGA---GAUGGGGAGAU .(..(((((.((((((..........((((((...(((.(((.......)))((((((.......))))))...)))))))))...------......))))))..---)))))..)... ( -34.81) >consensus AUUUCCAUCAGAAGUCAAAACCA_AACUCGCCAAACUCGUCGGGAAAUACGAUCCAUUUAUUCCUGGUGGAAAAGAGAGCGAGC_A_____AGGCAAGGAGUUUGAGA_GAUGGGGAGAU .(..(((((.((.((..........))))..(((((((.(((.......)))((((((.......))))))...........................)))))))....)))))..)... (-18.02 = -18.90 + 0.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:06 2006