
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,687,361 – 26,687,628 |
| Length | 267 |
| Max. P | 0.996670 |

| Location | 26,687,361 – 26,687,473 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -31.94 |
| Consensus MFE | -29.50 |
| Energy contribution | -30.34 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26687361 112 + 27905053 --------UGCGCUUUGGUCUCUGACCGAGACCCGUAAUAAUUGAAACCAAUUAGCUUGGAAAAACCUUUUAAUUGUGUGUUCAAUGCCCGGUCUUAAGUAAAUAUUUUCAUGGGCGCAG --------(((((((.((((...))))((((((.(.....(((((((((((((((...((.....))..))))))).)).))))))..).))))))................))))))). ( -31.00) >DroSec_CAF1 278115 120 + 1 UCGGAUUCUGCGCUUUGGUCUCUGACCGAGACCCGUAAUAAUUGAAACCAAUUAGCUUGGAAAAACCUUUUAAUUGUGUGUUCAAUGCCCGGUCUUAAGUAAAUAUUUUCAUGGGCGCAG .......((((((((.((((...))))((((((.(.....(((((((((((((((...((.....))..))))))).)).))))))..).))))))................)))))))) ( -32.90) >DroSim_CAF1 274872 120 + 1 UCGGAUUCUGCGCUUUGGUCUCUGACCGAGACCCGUUAUAAUUGAAACCAAUUAGCUUGGAAAAACCUUUUAAUAGUGUGUUCAAUGCCCGGUCUUAAGUAAAUAUUUUCAUGGGCGCAG .......((((((((.((((...))))((((((.....((((((....))))))(((((((...((((......)).)).))))).))..))))))................)))))))) ( -32.10) >DroEre_CAF1 294069 120 + 1 GCGGAUUCUGCGCUUUGGUCUCUGGCCGAGACCCGUAAUAAUUGAAACCAAUUAGCUUGGAAAAACCUUUUAAUUGUGUGCUCAAUGCCCGGUCUUAAGUAAAUAUUUUCAUGGGCGCAG .......((((((((.(((.....)))((((((.(.....(((((.(((((((((...((.....))..))))))).))..)))))..).))))))................)))))))) ( -32.50) >DroYak_CAF1 283078 120 + 1 GCGGAUUCUGCGCUUUGGUGUCUGACCGAGACCCGUAAUAAUUGAAACCAAUUAGCUUGGAAAAACCUUUUAAUUGUGUGCUCAAUGCCUGGUCUUAAGUAAAUAUUUUCAUGGGCGCAG .......((((((((.(((.....)))((((((.(.....(((((.(((((((((...((.....))..))))))).))..)))))..).))))))................)))))))) ( -31.20) >consensus GCGGAUUCUGCGCUUUGGUCUCUGACCGAGACCCGUAAUAAUUGAAACCAAUUAGCUUGGAAAAACCUUUUAAUUGUGUGUUCAAUGCCCGGUCUUAAGUAAAUAUUUUCAUGGGCGCAG .......((((((((.((((...))))((((((.(.....(((((((((((((((...((.....))..))))))).)).))))))..).))))))................)))))))) (-29.50 = -30.34 + 0.84)



| Location | 26,687,361 – 26,687,473 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.83 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -29.24 |
| Energy contribution | -29.84 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.73 |
| SVM RNA-class probability | 0.996670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26687361 112 - 27905053 CUGCGCCCAUGAAAAUAUUUACUUAAGACCGGGCAUUGAACACACAAUUAAAAGGUUUUUCCAAGCUAAUUGGUUUCAAUUAUUACGGGUCUCGGUCAGAGACCAAAGCGCA-------- ..((((...(((((..........((((((....((((......)))).....)))))).((((.....))))))))).........((((((.....))))))...)))).-------- ( -28.70) >DroSec_CAF1 278115 120 - 1 CUGCGCCCAUGAAAAUAUUUACUUAAGACCGGGCAUUGAACACACAAUUAAAAGGUUUUUCCAAGCUAAUUGGUUUCAAUUAUUACGGGUCUCGGUCAGAGACCAAAGCGCAGAAUCCGA ((((((...(((((..........((((((....((((......)))).....)))))).((((.....))))))))).........((((((.....))))))...))))))....... ( -32.30) >DroSim_CAF1 274872 120 - 1 CUGCGCCCAUGAAAAUAUUUACUUAAGACCGGGCAUUGAACACACUAUUAAAAGGUUUUUCCAAGCUAAUUGGUUUCAAUUAUAACGGGUCUCGGUCAGAGACCAAAGCGCAGAAUCCGA ((((((...(((((..........((((((.......................)))))).((((.....))))))))).........((((((.....))))))...))))))....... ( -29.80) >DroEre_CAF1 294069 120 - 1 CUGCGCCCAUGAAAAUAUUUACUUAAGACCGGGCAUUGAGCACACAAUUAAAAGGUUUUUCCAAGCUAAUUGGUUUCAAUUAUUACGGGUCUCGGCCAGAGACCAAAGCGCAGAAUCCGC ((((((...(((((..........((((((....((((......)))).....)))))).((((.....))))))))).........((((((.....))))))...))))))....... ( -32.30) >DroYak_CAF1 283078 120 - 1 CUGCGCCCAUGAAAAUAUUUACUUAAGACCAGGCAUUGAGCACACAAUUAAAAGGUUUUUCCAAGCUAAUUGGUUUCAAUUAUUACGGGUCUCGGUCAGACACCAAAGCGCAGAAUCCGC ((((((...................(((((.(..((((((.((.((((((...((.....))....)))))))))))))).....).))))).(((.....)))...))))))....... ( -27.50) >consensus CUGCGCCCAUGAAAAUAUUUACUUAAGACCGGGCAUUGAACACACAAUUAAAAGGUUUUUCCAAGCUAAUUGGUUUCAAUUAUUACGGGUCUCGGUCAGAGACCAAAGCGCAGAAUCCGA ((((((...(((((..........((((((....((((......)))).....)))))).((((.....))))))))).........((((((.....))))))...))))))....... (-29.24 = -29.84 + 0.60)



| Location | 26,687,473 – 26,687,593 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -26.22 |
| Energy contribution | -27.26 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26687473 120 + 27905053 GACAAUGCGAGGAUGUGAGAACAUGGCAAUGACCAAAAUAAUCCUGCUCCGUCCGUAGGAUAUAUUGUAGUGAAUGUUACCAGCGAAAGCGGUGCAAAGAAAUUGCCAUAUAAUUGCCAG ............((((....))))((((((...........((((((.......))))))(((((.(((((...(((.(((.((....)))))))).....))))).))))))))))).. ( -32.50) >DroSec_CAF1 278235 120 + 1 GACAAUGCGAGGAUGUGAGAACAUGGCAAUGACCAAAAUAAUCCUGCUCCGUUCGUAGCAUAUAUUGUAGUGAAUGUUACCAGCGAAAGCGGUCUAAAGAAAUUGCCAAAUAAUUGCCAG ......((((..((((....))))((((((((((...............((((((..(((.....)))..))))))......((....)))))).......))))))......))))... ( -29.51) >DroSim_CAF1 274992 120 + 1 GACAAUGCGAGGAUGUGAGAACAUGGCAAUGACCAAAAUAAUCCUGCUCCGUUCGUAGGAUAUAUUGUAGUGAAUGUUACCAGCGAAAGCGGUCUAAAGAAAUUGCCAAAUAAUUGCCAG ......((((..((((....))))((((((((((..(((((((((((.......))))))).))))................((....)))))).......))))))......))))... ( -31.11) >DroEre_CAF1 294189 120 + 1 GACAAUGCGAGGAUGUGAGAACAUGGCAAUGACCAAAAUAAUCCUGCUCCGUCCGCUGGAUAUGUUGUAGUGAAUGUUGCCAGCGAAAGCGGUAAGAAGAAAUUGCCAUAUAAUUGCCAG (((...((.(((((((....)).(((......))).....)))))))...)))..((((.(((((.(((((...(.(((((.((....))))))).)....))))).)))))....)))) ( -32.70) >DroYak_CAF1 283198 120 + 1 GACAAUGCGAGGAUGUGAGAACAUGGCAAUGACCAAAAUAAUCCUGCUCCGUACGCAGCAUAUAUGGUAGUGAAUGCUAACAGCGAAAGCGGUAAAAAGAAAUUGCCAUAUAAUUGCCAG ............((((....))))((((((.............((((.......))))..(((((((((((........((.((....)).))........))))))))))))))))).. ( -31.79) >consensus GACAAUGCGAGGAUGUGAGAACAUGGCAAUGACCAAAAUAAUCCUGCUCCGUCCGUAGGAUAUAUUGUAGUGAAUGUUACCAGCGAAAGCGGUAAAAAGAAAUUGCCAUAUAAUUGCCAG ............((((....))))((((((...........((((((.......))))))(((((.(((((......((((.((....)))))).......))))).))))))))))).. (-26.22 = -27.26 + 1.04)

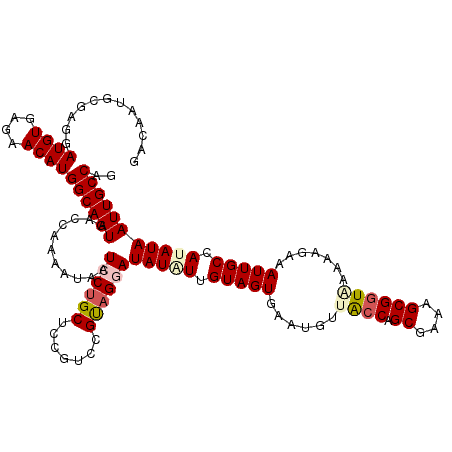
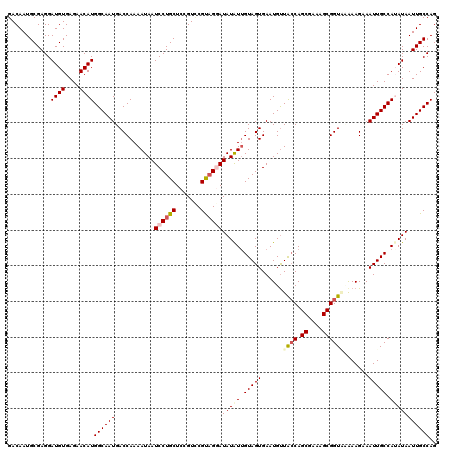
| Location | 26,687,513 – 26,687,628 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -31.45 |
| Consensus MFE | -20.38 |
| Energy contribution | -21.66 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627343 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26687513 115 + 27905053 AUCCUGCUCCGUCCGUAGGAUAUAUUGUAGUGAAUGUUACCAGCGAAAGCGGUGCAAAGAAAUUGCCAUAUAAUUGCCAGGAAAAAAAACCAAGACAAAGGCA-----GUGCAGAAUGGA ...(((((((.......)))(((((.(((((...(((.(((.((....)))))))).....))))).)))))((((((.....................))))-----))))))...... ( -28.00) >DroSec_CAF1 278275 113 + 1 AUCCUGCUCCGUUCGUAGCAUAUAUUGUAGUGAAUGUUACCAGCGAAAGCGGUCUAAAGAAAUUGCCAAAUAAUUGCCAGGAAAA--AAGCAAGACAAAGGCU-----CUGGAGAAUGGA .(((..((((.((((((((((.(((....))).)))))))..((....))........)))...(((......((((........--..))))......))).-----..))))...))) ( -29.00) >DroSim_CAF1 275032 118 + 1 AUCCUGCUCCGUUCGUAGGAUAUAUUGUAGUGAAUGUUACCAGCGAAAGCGGUCUAAAGAAAUUGCCAAAUAAUUGCCAGGAAAA--AAGCAAGACAAAGAAUAGAGAAUGGAGAAUGGA .(((..((((((((..........((((..((....((.((.((....))(((...((....)))))............)).)).--...))..))))........))))))))...))) ( -28.47) >DroEre_CAF1 294229 113 + 1 AUCCUGCUCCGUCCGCUGGAUAUGUUGUAGUGAAUGUUGCCAGCGAAAGCGGUAAGAAGAAAUUGCCAUAUAAUUGCCAGGAAAA--AAGCAAGACAAAGGCG-----GUGGAGAAUGGA .(((..(((((.(((((...(((((.(((((...(.(((((.((....))))))).)....))))).))))).((((........--..))))......))))-----))))))...))) ( -38.00) >DroYak_CAF1 283238 113 + 1 AUCCUGCUCCGUACGCAGCAUAUAUGGUAGUGAAUGCUAACAGCGAAAGCGGUAAAAAGAAAUUGCCAUAUAAUUGCCAGGAAAA--AAGCAAGACAAAGGCG-----GUGGAGAAUGGA .(((..((((...(((....(((((((((((........((.((....)).))........))))))))))).((((........--..)))).......)))-----..))))...))) ( -33.79) >consensus AUCCUGCUCCGUCCGUAGGAUAUAUUGUAGUGAAUGUUACCAGCGAAAGCGGUAAAAAGAAAUUGCCAUAUAAUUGCCAGGAAAA__AAGCAAGACAAAGGCG_____GUGGAGAAUGGA .(((..(((((.........(((((.(((((......((((.((....)))))).......))))).)))))...(((.....................))).......)))))...))) (-20.38 = -21.66 + 1.28)

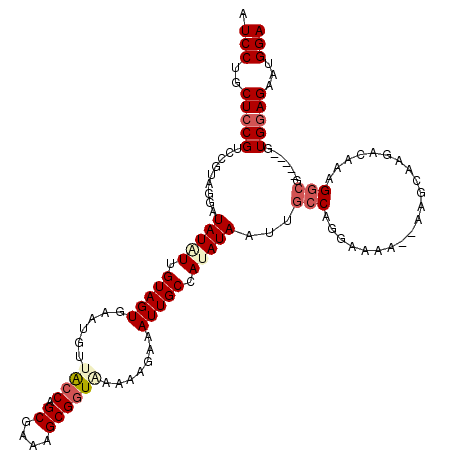
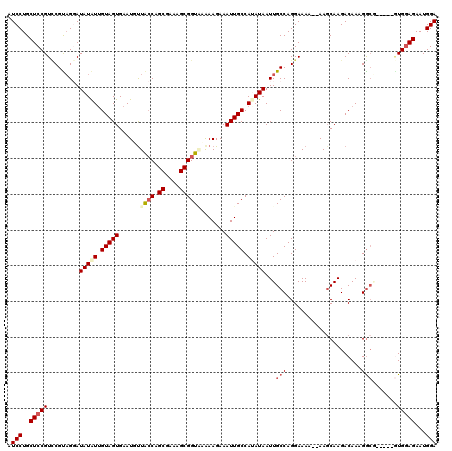
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:46:01 2006