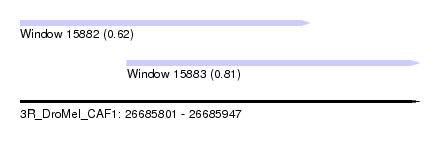
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,685,801 – 26,685,947 |
| Length | 146 |
| Max. P | 0.806080 |

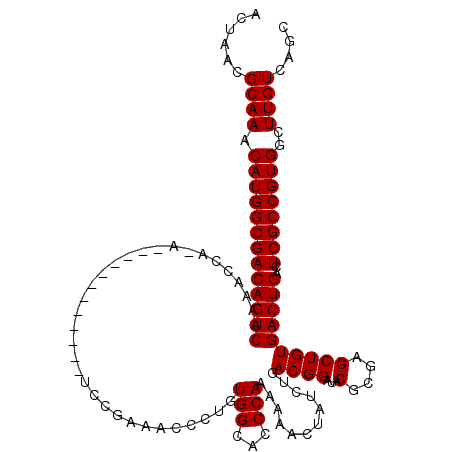
| Location | 26,685,801 – 26,685,907 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -23.60 |
| Energy contribution | -24.00 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.619558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26685801 106 + 27905053 AGCGGCUG-GCGACAGCGACAACCCGCUGCGGGGAUUAGCACUAACGCAAACAUGGCGACAGUCAAAACCA-A------------UCCGAAACCCUGUGGCACCCAAAAAAUUAUCUGAC ....((((-....))))........((..(((((....((......)).....((((....))))......-.------------.......)))))..))................... ( -29.20) >DroSec_CAF1 276561 106 + 1 AGCGGCUG-GCGACAGCGACAACCCGCUGCGGGGAUUAGCACUAACGCAAACAUGGCGACAGUCAAAACCA-A------------UCCGAAACCCUGUGGCACCCAAAAAACUAUCUGAC ....((((-....))))........((..(((((....((......)).....((((....))))......-.------------.......)))))..))................... ( -29.20) >DroSim_CAF1 273309 106 + 1 AGCGGCUG-GCGACAGCGACAACCCGCUGCGGGGAUUAGCACUAACGCAAACAUGGCGACAGUCAAAACCA-A------------UCCAAAACCCUGUGGCACCCAAAAAACUAUCUGAC ....((((-....))))........((..(((((....((......)).....((((....))))......-.------------.......)))))..))................... ( -29.20) >DroEre_CAF1 292506 108 + 1 AGCGGCUGGGCGACAGCGACAACCCGCUGCGGGGAUUAGCACUAACGCAAACAUGGCGACAGUCAAAACCAAA------------UCCGAAGCCCUAUGGCACCCAAAAAAGUAUCUGAC .((.(..(((((.(((((......))))))..(((((.((......)).....((((....))))......))------------)))...))))..).))................... ( -30.40) >DroYak_CAF1 281505 119 + 1 AGCGGCUG-GCGACAGCGACAACCCGCUGCGGGGAUUAGCACUAACGCAAACAUGGCGACAGUCAAAACCAAAUCCCAAUCCGAAUCCGAAGCCCUAUGGCACCCAAAAAAGUAUCUGAC ..(((.((-(((.(((((......)))))))((((((.((......)).....((((....))))......))))))...)))...)))..(((....)))................... ( -30.90) >consensus AGCGGCUG_GCGACAGCGACAACCCGCUGCGGGGAUUAGCACUAACGCAAACAUGGCGACAGUCAAAACCA_A____________UCCGAAACCCUGUGGCACCCAAAAAACUAUCUGAC ....((((.....))))........(((((((((....((......)).....((((....))))...........................)))))))))................... (-23.60 = -24.00 + 0.40)

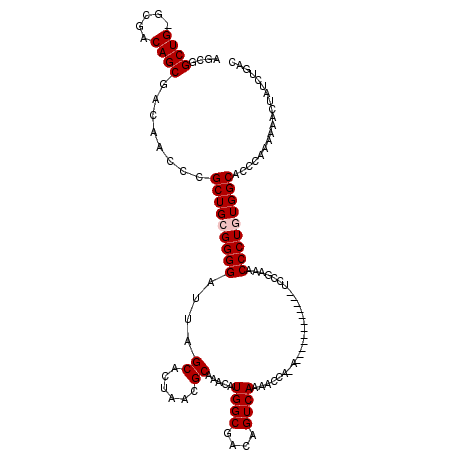
| Location | 26,685,840 – 26,685,947 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -30.05 |
| Consensus MFE | -25.44 |
| Energy contribution | -25.44 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26685840 107 + 27905053 ACUAACGCAAACAUGGCGACAGUCAAAACCA-A------------UCCGAAACCCUGUGGCACCCAAAAAAUUAUCUGACGGAUACGCGAGCUGUGACUGACUCGCCGUGGCUUGUCAGC ......((((.((((((((((((((......-(------------((((........(((...))).............))))).(....)...))))))..))))))))..)))).... ( -27.90) >DroSec_CAF1 276600 107 + 1 ACUAACGCAAACAUGGCGACAGUCAAAACCA-A------------UCCGAAACCCUGUGGCACCCAAAAAACUAUCUGACGGAUACGCGAGCUGUGACUGACUCGCCGUGGCUUGUCAGC ......((((.((((((((((((((......-(------------((((........(((...))).....(.....).))))).(....)...))))))..))))))))..)))).... ( -28.80) >DroSim_CAF1 273348 107 + 1 ACUAACGCAAACAUGGCGACAGUCAAAACCA-A------------UCCAAAACCCUGUGGCACCCAAAAAACUAUCUGACGGAUACGCGAGCUGUGACUGACUCGCCGUGGCUUGUCAGC ......((((.((((((((((((((......-(------------(((.........(((...))).....(.....)..)))).(....)...))))))..))))))))..)))).... ( -26.80) >DroEre_CAF1 292546 108 + 1 ACUAACGCAAACAUGGCGACAGUCAAAACCAAA------------UCCGAAGCCCUAUGGCACCCAAAAAAGUAUCUGACGGAUACGCGAGCUGUGACUGACUCGCCGUGGCUUGUCAGC ......((((.((((((((((((((....((..------------(((...(((....)))..........(((((.....)))))).))..))))))))..))))))))..)))).... ( -32.80) >DroYak_CAF1 281544 120 + 1 ACUAACGCAAACAUGGCGACAGUCAAAACCAAAUCCCAAUCCGAAUCCGAAGCCCUAUGGCACCCAAAAAAGUAUCUGACGGAUACGCGAGCUGUGACUGACUCGCCGUGGCUUGUCAGC ......((((.((((((((((((((..............((((.(((((..(((....)))((........))......))))).)).))....))))))..))))))))..)))).... ( -33.97) >consensus ACUAACGCAAACAUGGCGACAGUCAAAACCA_A____________UCCGAAACCCUGUGGCACCCAAAAAACUAUCUGACGGAUACGCGAGCUGUGACUGACUCGCCGUGGCUUGUCAGC ......((((.(((((((((((((.................................(((...)))............((((...(....))))))))))..))))))))..)))).... (-25.44 = -25.44 + -0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:45:54 2006