| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,682,513 – 26,682,815 |
| Length | 302 |
| Max. P | 0.959609 |

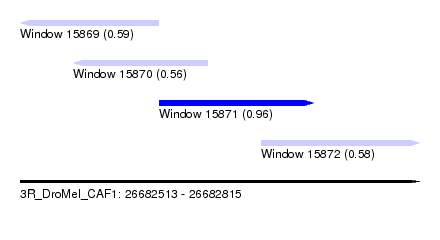
| Location | 26,682,513 – 26,682,618 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.34 |
| Mean single sequence MFE | -25.94 |
| Consensus MFE | -23.88 |
| Energy contribution | -23.92 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26682513 105 - 27905053 --------------AACCAGGCUUAGUUGGCAGCGCA-UAAGCCACAGAAACAAUUAUAUACCCGCUUAUCUGAGCGGCAGAUAUAGAAAAUUGAUCUGGCGAUAAAUCGAGCUACUCAA --------------.....((((..(((....((((.-...))..((((..(((((((((..((((((....))))))...)))))....)))).))))))....)))..))))...... ( -26.00) >DroSec_CAF1 273191 105 - 1 --------------UACCAGGCUUAGUUGGCAGCGCA-UAAGCCACAGAAACAAUUAUAUACCCGCUUAUCUGGGCGGCAGAUAUAGAAAAUUGAUCUGGCGAUAAAUCGAGCUACUCAA --------------.....((((..(((....((((.-...))..((((..(((((((((..((((((....))))))...)))))....)))).))))))....)))..))))...... ( -25.20) >DroSim_CAF1 269931 105 - 1 --------------AACCAGGCUUAGUUGGCAGCGCA-UAAGCCACAGAAACAAUUAUAUACCCGCUUAUCUGGGCGGCAGAUAUAGAAAAUUGAUCUGGCGAUAAAUCGAGCUACUCAA --------------.....((((..(((....((((.-...))..((((..(((((((((..((((((....))))))...)))))....)))).))))))....)))..))))...... ( -25.20) >DroEre_CAF1 289098 105 - 1 --------------AACCAGGCUUAGUCGGCAGCGCA-UAAGCCACAGUAACAAUUAUAUACCCGCUUAUCUGGGCGGCAGAUAUAGAAAAUUGAUCUGGCGAUAAAUCGAGCUACUCAA --------------.....((((((..((....))..-))))))..........((((((..((((((....))))))...))))))....((((..((((..........)))).)))) ( -24.10) >DroYak_CAF1 278089 120 - 1 GAAAAGCAGAAAAGCAGCAGGCUUAGUUGGCAGCGCAGUAAGCCACAGUAACAAUUAUAUACCCGCUUAUCUGGGCGGCAGAUAUAGAAAAUUGAUCUGGCGAUAAAUCGAGCUACUCAA ((..(((.((......((.((((((.(((......))))))))).(((...(((((((((..((((((....))))))...)))))....))))..)))))......))..)))..)).. ( -29.20) >consensus ______________AACCAGGCUUAGUUGGCAGCGCA_UAAGCCACAGAAACAAUUAUAUACCCGCUUAUCUGGGCGGCAGAUAUAGAAAAUUGAUCUGGCGAUAAAUCGAGCUACUCAA ...................((((..(((....((((.....))..(((...(((((((((..((((((....))))))...)))))....))))..)))))....)))..))))...... (-23.88 = -23.92 + 0.04)



| Location | 26,682,553 – 26,682,655 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 95.92 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -25.62 |
| Energy contribution | -25.25 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26682553 102 - 27905053 GGCUCAGGGCAAGAAGCGAAACGAAAACCAAGACCAAAACCAGGCUUAGUUGGCAGCGCAUAAGCCACAGAAACAAUUAUAUACCCGCUUAUCUGAGCGGCA .(((((((.....(((((.......(((.(((.((.......))))).)))(((.........)))...................))))).))))))).... ( -25.20) >DroSec_CAF1 273231 102 - 1 GGCUCAGAGCAAGAAGCGAAACGGAAACCAGGACCAAUACCAGGCUUAGUUGGCAGCGCAUAAGCCACAGAAACAAUUAUAUACCCGCUUAUCUGGGCGGCA .(((((((.....(((((....(....)..............((((((....((...)).))))))...................))))).))))))).... ( -27.40) >DroSim_CAF1 269971 102 - 1 GGCUCAGAGCAAGAAGCGAAACGGAAACCAGGACCAAAACCAGGCUUAGUUGGCAGCGCAUAAGCCACAGAAACAAUUAUAUACCCGCUUAUCUGGGCGGCA .(((((((.....(((((....(....)..............((((((....((...)).))))))...................))))).))))))).... ( -27.40) >DroEre_CAF1 289138 102 - 1 GGCUCAGAGCAAGAAGCGAAGCGAAAACCAGGACCAAAACCAGGCUUAGUCGGCAGCGCAUAAGCCACAGUAACAAUUAUAUACCCGCUUAUCUGGGCGGCA .(((((((((.....)).(((((.......((.......)).((((((..((....))..))))))...................))))).))))))).... ( -28.50) >consensus GGCUCAGAGCAAGAAGCGAAACGAAAACCAGGACCAAAACCAGGCUUAGUUGGCAGCGCAUAAGCCACAGAAACAAUUAUAUACCCGCUUAUCUGGGCGGCA .(((((((.....(((((....(....)..............((((((....((...)).))))))...................))))).))))))).... (-25.62 = -25.25 + -0.38)



| Location | 26,682,618 – 26,682,735 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.16 |
| Mean single sequence MFE | -30.35 |
| Consensus MFE | -26.98 |
| Energy contribution | -26.58 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959609 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26682618 117 + 27905053 ---UUGGUCUUGGUUUUCGUUUCGCUUCUUGCCCUGAGCCUUACUUAGUAGCUUAAGUUCUGCUUAACUUACUUUAGUGGGCAAAUGGCAAAUUUUAGGCGGAAAAUUCUACGAAAUUCU ---........(..(((((((((((((.(((((.(..((((.(((.(((((.(((((.....))))).)))))..)))))))..).))))).....))))))).......))))))..). ( -31.71) >DroSec_CAF1 273296 117 + 1 ---UUGGUCCUGGUUUCCGUUUCGCUUCUUGCUCUGAGCCUUACUUAGUACCUUAAGUUCUGCUUAACUUACUUUAGUGGGCAAAUGGCAAAUUUUCGGCGGAAAAUUCUACGAAGUUCU ---((.((.....((((((((.......(((((.(..((((.(((.((((..(((((.....)))))..))))..)))))))..).)))))......)))))))).....)).))..... ( -28.42) >DroSim_CAF1 270036 117 + 1 ---UUGGUCCUGGUUUCCGUUUCGCUUCUUGCUCUGAGCCUUACUUAGUACCUUAAGUUCUGCUUAACUUACUUUAGUGGGCAAAUGGCAAAUUUUCGGCGGAAAAUUCUACGAAGUUCU ---((.((.....((((((((.......(((((.(..((((.(((.((((..(((((.....)))))..))))..)))))))..).)))))......)))))))).....)).))..... ( -28.42) >DroEre_CAF1 289203 117 + 1 ---UUGGUCCUGGUUUUCGCUUCGCUUCUUGCUCUGAGCCUUACUUAGUUCCUUAAGUUCUGCUUAACUUACUUUAGUGGGCAAAUGGCAAAUUUGAGGCGGAAAAUUCUACGAAGUUCU ---((.((.....(((((((((((....(((((.(..((((.(((.(((...(((((.....)))))...)))..)))))))..).)))))...))))))))))).....)).))..... ( -31.20) >DroYak_CAF1 278209 120 + 1 UGGUUGCUCCUGGUUUUCGUUUCGCUUCUUGUUCUGAGCCUUACUUAGUGCCUUAAGUUCUGCUUAACUUACUUUAGUGGGCAAAUGGCAAAUUUGAGGCGGAGAAUUCUACGAAGUUCU .((.....)).(..((((((....((((.....(((((.....))))).((((((((((.((((...(((((....))))).....))))))))))))))))))......))))))..). ( -32.00) >consensus ___UUGGUCCUGGUUUUCGUUUCGCUUCUUGCUCUGAGCCUUACUUAGUACCUUAAGUUCUGCUUAACUUACUUUAGUGGGCAAAUGGCAAAUUUUAGGCGGAAAAUUCUACGAAGUUCU .......((.((((((((((((......(((((.(..((((.(((.((((..(((((.....)))))..))))..)))))))..).))))).....)))))))))...))).))...... (-26.98 = -26.58 + -0.40)



| Location | 26,682,695 – 26,682,815 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -28.08 |
| Energy contribution | -28.08 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26682695 120 + 27905053 GCAAAUGGCAAAUUUUAGGCGGAAAAUUCUACGAAAUUCUUUGUAUUUGCAUACACAUCCGUGGCGGCAUUGUCUUUGACGCUUAGUCGCAUAAUUUGCCAGACAAAACGGUCAUAGGAC (((((((.((((.(((..(..((....))..).)))...))))))))))).......((((((((.(..((((((..(((.....)))(((.....))).))))))..).))))).))). ( -29.80) >DroSec_CAF1 273373 120 + 1 GCAAAUGGCAAAUUUUCGGCGGAAAAUUCUACGAAGUUCUUUGUAUUUGCAUACACAUCCGUGGCGGCAUUGUCUUUGACGCUUAGUCGCAUAAUUUGCCAGACAAAACAGUCAUAGGAC (((((((.((((.(((((..(((....))).)))))...))))))))))).......((((((((....((((((..(((.....)))(((.....))).))))))....))))).))). ( -33.60) >DroSim_CAF1 270113 120 + 1 GCAAAUGGCAAAUUUUCGGCGGAAAAUUCUACGAAGUUCUUUGUAUUUGCAUACACAUCCGUGGCGGCAUUGUCUUUGACGCUUAGUCGCAUAAUUUGCCAGACAAAACGGUCAUAGGAC (((((((.((((.(((((..(((....))).)))))...))))))))))).......((((((((.(..((((((..(((.....)))(((.....))).))))))..).))))).))). ( -35.00) >DroEre_CAF1 289280 120 + 1 GCAAAUGGCAAAUUUGAGGCGGAAAAUUCUACGAAGUUCUUUGUAUUUGCAUACACAUCCGUGGCGGCAUUGUCUUUGACGCUUAGUCGCAUAAUUUGCCAGACAAAACGGUCAGCGGAG ((...(((((((((.(((((((.......((((((....))))))..(((...(((....)))...)))))))))).(((.....)))....)))))))))(((......))).)).... ( -32.50) >DroYak_CAF1 278289 120 + 1 GCAAAUGGCAAAUUUGAGGCGGAGAAUUCUACGAAGUUCUUUGUAUUUGCAUACACAGCCGUGGCGGCAUUGUCUUUGCCGCUUAGUCGCAUAAUUUGCCAGACAAAACGGUCAGCGGAG ((...(((((((((....((((((((((......))))))))))...(((...(((....)))((((((.......))))))......))).)))))))))(((......))).)).... ( -40.10) >consensus GCAAAUGGCAAAUUUUAGGCGGAAAAUUCUACGAAGUUCUUUGUAUUUGCAUACACAUCCGUGGCGGCAUUGUCUUUGACGCUUAGUCGCAUAAUUUGCCAGACAAAACGGUCAUAGGAC .....(((((((((....((((.......((((((....))))))..(((...(((....)))...))).................))))..)))))))))(((......)))....... (-28.08 = -28.08 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:45:44 2006