
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,682,223 – 26,682,375 |
| Length | 152 |
| Max. P | 0.999992 |

| Location | 26,682,223 – 26,682,335 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 96.34 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -26.69 |
| Energy contribution | -26.89 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26682223 112 + 27905053 UAGAUUUUAAUGGGCAGGAUUACCCUUUUGCUAAAUA-------CCAAAAUCCUGGACGGGCGGUCUGCGCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAA ............(((((((((......((....))..-------....))))))(((((((.((.(((((...))))))))))..(((((((.......)))))))))))....))).. ( -25.40) >DroSec_CAF1 272909 112 + 1 UAGAUUUUAAUGGGCAGGAUUACCCUUUUGCUAAAUA-------CCAAAAUCCUGGCCGGGCGGUCUGCGCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAA ..........(((.(((((((......((....))..-------....))))))).)))(((((.(((((...)))))..))...(((((((.......)))))))........))).. ( -29.60) >DroSim_CAF1 269649 112 + 1 UAGAUUUUAAUGGGCAGGAUUACCCUUUUGCUAAAUA-------CCAAAAUCCUGGCCGGGCGGUCUGCGCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAA ..........(((.(((((((......((....))..-------....))))))).)))(((((.(((((...)))))..))...(((((((.......)))))))........))).. ( -29.60) >DroEre_CAF1 288804 112 + 1 UAGAUUUUAAUGGCCAGGAUUACCCUUUUGCUAAAUA-------CCAAAAUCCUGGCCGGGCGGUCUGCUCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAA ..........(((((((((((......((....))..-------....)))))))))))(((((.((((.....))))..))...(((((((.......)))))))........))).. ( -33.30) >DroYak_CAF1 277793 119 + 1 UAGAUUUUAAUGGGCAGGAUUACCCUUUUGCUAAAUACCAAAUACCAAAAUCCUGGCCGGGCGGUCUGCUCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAA ..........(((.(((((((.....((((........))))......))))))).)))(((((.((((.....))))..))...(((((((.......)))))))........))).. ( -30.60) >consensus UAGAUUUUAAUGGGCAGGAUUACCCUUUUGCUAAAUA_______CCAAAAUCCUGGCCGGGCGGUCUGCGCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAA ..........(((.(((((((...........................))))))).)))(((((.((((.....))))..))...(((((((.......)))))))........))).. (-26.69 = -26.89 + 0.20)


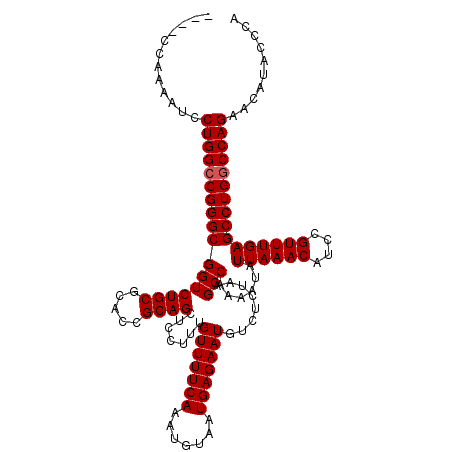
| Location | 26,682,260 – 26,682,375 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 95.37 |
| Mean single sequence MFE | -35.38 |
| Consensus MFE | -35.08 |
| Energy contribution | -35.28 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.71 |
| SVM RNA-class probability | 0.999992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26682260 115 + 27905053 ----CCAAAAUCCUGGACGGGCGGUCUGCGCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAAAAUAUUAAACAUCCGUUUGAGCCUGGCCAGAACAUACCCA ----........((((.(((((((((((((...))))).......(((((((.......)))))))........)))......((((((....))))))))))).)))).......... ( -30.20) >DroSec_CAF1 272946 115 + 1 ----CCAAAAUCCUGGCCGGGCGGUCUGCGCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAAAAUAUUAAACAUCCGUUUGAGCCUGGCCAGAACACAACCA ----........((((((((((((((((((...))))).......(((((((.......)))))))........)))......((((((....)))))))))))))))).......... ( -37.30) >DroSim_CAF1 269686 115 + 1 ----CCAAAAUCCUGGCCGGGCGGUCUGCGCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAAAAUAUUAAACAUCCGUUUGAGCCUGGCCAGAACACAACCA ----........((((((((((((((((((...))))).......(((((((.......)))))))........)))......((((((....)))))))))))))))).......... ( -37.30) >DroEre_CAF1 288841 115 + 1 ----CCAAAAUCCUGGCCGGGCGGUCUGCUCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAAAAUAUUAAACAUCCGUUUGAGCCUGGCCAGCACAUACCCA ----........(((((((((((((((((.....)))).......(((((((.......)))))))........)))......((((((....)))))))))))))))).......... ( -35.70) >DroYak_CAF1 277833 119 + 1 AAUACCAAAAUCCUGGCCGGGCGGUCUGCUCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAAAAUAUUAAACAUCCGUUUGAGCCUGGCCAGAACAUGCAUA ............(((((((((((((((((.....)))).......(((((((.......)))))))........)))......((((((....)))))))))))))))).......... ( -36.40) >consensus ____CCAAAAUCCUGGCCGGGCGGUCUGCGCACCGCAGCUCCUUUGUUUUCAAAUGUAAUGAGAAUGUCUCUAAGCCAAAAUAUUAAACAUCCGUUUGAGCCUGGCCAGAACAUACCCA ............(((((((((((((((((.....)))).......(((((((.......)))))))........)))......((((((....)))))))))))))))).......... (-35.08 = -35.28 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:45:40 2006