
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,634,450 – 26,634,584 |
| Length | 134 |
| Max. P | 0.842338 |

| Location | 26,634,450 – 26,634,544 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.74 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -23.21 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.86 |
| SVM decision value | -0.05 |
| SVM RNA-class probability | 0.508683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26634450 94 + 27905053 AUUCGAAGGAAGUGAGUAGUGAGUGGCGAAAAGCGGAGACGCACUGCCGCGCUGGAAAGGCGGAGUGAAUUGAGUGCAUGCGAAUGCGUGUGGA ((((((..........................(((....)))(((.((((.((....)))))))))...))))))((((((....))))))... ( -28.90) >DroSec_CAF1 226115 93 + 1 AUUCGGAGGAAGUGAGUAGUGAGUGGCGAAAAG-GGAGACGCACUUCCGCGCUGGAAAGGCGGAGUGAAUCGAGUGCAUGCGAAUGGGUGUGGA ((((((.(((((((.((........))......-(....).))))))).((((.....)))).......)))))).(((((......))))).. ( -25.00) >DroSim_CAF1 221873 94 + 1 AUUCGGAGGAAGUGAGUUGUGAGUGGCGAAAAGCGGAGACGCACUGCCGCGCUGGAAAGGCGGAGUGAAUCGAGUGCAUGCGAAUGGGUGUGGA ((((((..........((((.....))))...(((....)))(((.((((.((....)))))))))...)))))).(((((......))))).. ( -26.80) >consensus AUUCGGAGGAAGUGAGUAGUGAGUGGCGAAAAGCGGAGACGCACUGCCGCGCUGGAAAGGCGGAGUGAAUCGAGUGCAUGCGAAUGGGUGUGGA ((((((..........................(((....)))(((.((((.((....)))))))))...)))))).(((((......))))).. (-23.21 = -24.60 + 1.39)



| Location | 26,634,482 – 26,634,584 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -16.76 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26634482 102 + 27905053 G--CGG-----------AGACG----C-ACUGCCGCGCUGGAAAGGCGGAGUGAAUUGAGUGCAUGCGAAUGCGUGUGGAGAGCAAAUCGAAGUGCAAGUGCGUGCCAGCGAAUAAGAAG (--((.-----------...))----)-(((.((((.((....)))))))))...(((...(((((((..((((..(.((.......)).)..))))..)))))))...)))........ ( -37.30) >DroSec_CAF1 226147 101 + 1 G---GG-----------AGACG----C-ACUUCCGCGCUGGAAAGGCGGAGUGAAUCGAGUGCAUGCGAAUGGGUGUGGAUAGCAAAUCGAAGUGCAAGUGCGGGCCAGCGAAUAAGAGG .---((-----------...((----(-((((((((.((....)))))))(((..((((((.(((((......))))).))......))))..))).))))))..))............. ( -32.00) >DroSim_CAF1 221905 102 + 1 G--CGG-----------AGACG----C-ACUGCCGCGCUGGAAAGGCGGAGUGAAUCGAGUGCAUGCGAAUGGGUGUGGAUAGCAAAGCGAAGUGCAAGUGCGUGCCAGCGAAUAAGAAG (--((.-----------...))----)-(((.((((.((....)))))))))...(((...(((((((.....(..(.....((...))...)..)...)))))))...)))........ ( -33.90) >DroEre_CAF1 235505 108 + 1 GUGCGG-----------UGGCAGUUGCGGCUGCAAAGC-GGAAAAGCAGAGUGAAUAAAGUGCAUGCGAAUGGGCGUGGAAAGCGAAACGAAGUGCAAGUGCGUGCCAGCGAAUAAGAAG ...((.-----------(((((.((((..((((...))-))....))))..........(..(.((((..(.(.(((.....)))...).)..)))).)..).))))).))......... ( -30.20) >DroYak_CAF1 229406 119 + 1 GUGCGGCUGAAAGGAAGUGCAAGUUGCAAGUGGAGUGC-AGCAAAGCAGAGUAAAUAAAGUGCAUGCGAAUGGGUGUGGAAAGCAAAACGAAGUGCAAGUGCGUGCCAGCGAAUAAGAAG .((((((..........(((..((((((.......)))-)))...)))...........(..(.((((..(.(.(((.....)))...).)..)))).)..)..))).)))......... ( -26.70) >consensus G__CGG___________AGACG____C_ACUGCCGCGCUGGAAAGGCGGAGUGAAUAGAGUGCAUGCGAAUGGGUGUGGAAAGCAAAACGAAGUGCAAGUGCGUGCCAGCGAAUAAGAAG .............................((....((((((....(((..............(((((......)))))....(((........)))...)))...))))))....))... (-16.76 = -17.08 + 0.32)

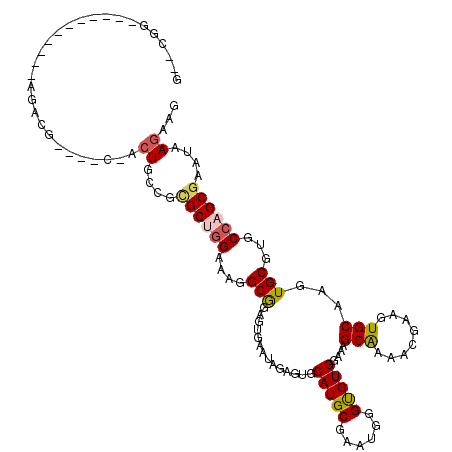

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:45:27 2006