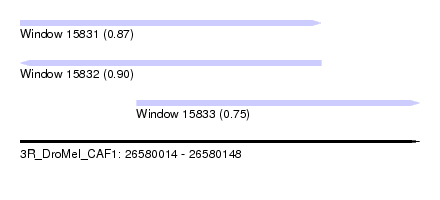
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,580,014 – 26,580,148 |
| Length | 134 |
| Max. P | 0.898882 |

| Location | 26,580,014 – 26,580,115 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -15.77 |
| Energy contribution | -15.39 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
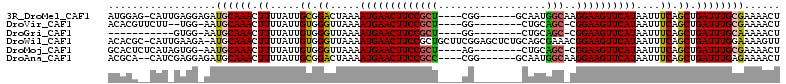
>3R_DroMel_CAF1 26580014 101 + 27905053 AUGGAG-CAUUGAGGAGAUGCAAACUUUUAUUGCGGACUAAAAUGAACUUCCGCU----CGG------GCAAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU .....(-((((.....))))).........(((((((.....(((((((((((((----...------.....)))..))))))))))....((....)))))))))..... ( -24.50) >DroVir_CAF1 209476 96 + 1 ACACGUUCUU--UGG-AAUGCAAACUUUUAUUGUGGGUUAAAAUGAACUUCCGCU----GG--------CUGCAGC-CGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU ...(((((..--..)-))))..(((((.......)))))...(((((((((((((----(.--------...))).-)))))))))))..((((.((......))))))... ( -27.60) >DroGri_CAF1 339761 87 + 1 -----------GUGG-AAUGCAAACUUUUAUUGUGGGUUAAAAUGAACUUCCGCU----GG--------CUGCAGC-CGGAAGUUCAUAAUUUCAGCUGAUUUGCAAAAACU -----------(..(-(..((.(((((.......)))))...(((((((((((((----(.--------...))).-))))))))))).......))...))..)....... ( -26.30) >DroWil_CAF1 195149 110 + 1 ACACGC-CAUUGAAGA-AUGCAAACUUUUAUUGUGGGUUAAAAUGAACUUCCGCUGCUUCGGAGCUCUGCAGCGAAACGGAAGUUCAUAAUUUCAGCUGAUUUGGAAAAGUU ....((-.(((....)-)))).(((((((...((.((((((((((((((((((.((((.(((....))).))))...)))))))))))..))).)))).))....))))))) ( -30.90) >DroMoj_CAF1 218300 98 + 1 GCACUCUCAUAGUGG-AAUGCAAACUUUUAUUGUGGGUUAAAAUGAACUUCCGCU----AG--------CUGCAGC-CGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU .....((((((((((-((.......)))))))))))).....(((((((((((((----..--------....)).-)))))))))))..((((.((......))))))... ( -29.20) >DroAna_CAF1 170553 100 + 1 ACGCA--CAUCGAGGAGAUGCAAACUUUUAUUGCGGACUAAAAUGAACUUCCGCC----CGG------GCAAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGAGAAAACU .(.((--.((((..(((((((((.......))))........(((((((((((((----...------.....)))..)))))))))).)))))...)))).)).)...... ( -25.90) >consensus ACACA__CAU_GAGG_AAUGCAAACUUUUAUUGUGGGUUAAAAUGAACUUCCGCU____CG________CAGCAGC_CGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU ..................((((((.((.....((.((.....(((((((((((((..................)))..))))))))))....)).)).))))))))...... (-15.77 = -15.39 + -0.39)

| Location | 26,580,014 – 26,580,115 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -14.29 |
| Energy contribution | -14.82 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.898882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26580014 101 - 27905053 AGUUUUCGCAAAUCAGCUGAAAUUAUGAACUUCCUUGCCAUUGC------CCG----AGCGGAAGUUCAUUUUAGUCCGCAAUAAAAGUUUGCAUCUCCUCAAUG-CUCCAU .......(((.....((((((...((((((((((..(((.....------..)----.))))))))))))))))))..((((.......))))..........))-)..... ( -23.20) >DroVir_CAF1 209476 96 - 1 AGUUUUCGCAAAUCAGCUGAAAUUAUGAACUUCCG-GCUGCAG--------CC----AGCGGAAGUUCAUUUUAACCCACAAUAAAAGUUUGCAUU-CCA--AAGAACGUGU .......((((((............((((((((((-.(((...--------.)----))))))))))))(((((........)))))))))))...-...--.......... ( -23.30) >DroGri_CAF1 339761 87 - 1 AGUUUUUGCAAAUCAGCUGAAAUUAUGAACUUCCG-GCUGCAG--------CC----AGCGGAAGUUCAUUUUAACCCACAAUAAAAGUUUGCAUU-CCAC----------- ......(((((((............((((((((((-.(((...--------.)----))))))))))))(((((........))))))))))))..-....----------- ( -23.70) >DroWil_CAF1 195149 110 - 1 AACUUUUCCAAAUCAGCUGAAAUUAUGAACUUCCGUUUCGCUGCAGAGCUCCGAAGCAGCGGAAGUUCAUUUUAACCCACAAUAAAAGUUUGCAU-UCUUCAAUG-GCGUGU (((((((.....((....))....((((((((((((((((..((...))..))))...)))))))))))).............))))))).((((-.((.....)-).)))) ( -24.80) >DroMoj_CAF1 218300 98 - 1 AGUUUUCGCAAAUCAGCUGAAAUUAUGAACUUCCG-GCUGCAG--------CU----AGCGGAAGUUCAUUUUAACCCACAAUAAAAGUUUGCAUU-CCACUAUGAGAGUGC ((((((.((......)).))))))(((((((((((-.(((...--------.)----))))))))))))).....................(((((-((.....).)))))) ( -26.20) >DroAna_CAF1 170553 100 - 1 AGUUUUCUCAAAUCAGCUGAAAUUAUGAACUUCCUUGCCAUUGC------CCG----GGCGGAAGUUCAUUUUAGUCCGCAAUAAAAGUUUGCAUCUCCUCGAUG--UGCGU ...............((.((....((((((((((..(((.....------...----))))))))))))).....)).)).......((..(((((.....))))--))).. ( -26.40) >consensus AGUUUUCGCAAAUCAGCUGAAAUUAUGAACUUCCG_GCCGCAG________CG____AGCGGAAGUUCAUUUUAACCCACAAUAAAAGUUUGCAUU_CCAC_AUG__CGUGU .......((((((............(((((((((..(((..................))))))))))))(((((........)))))))))))................... (-14.29 = -14.82 + 0.53)

| Location | 26,580,053 – 26,580,148 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -21.07 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.752523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26580053 95 + 27905053 AAAUGAACUUCCGCUCGGGCAAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU-UUCGCCACAUUCGCU-GCACCCCCUCCACGAUCC ..(((((((((((((........)))..)))))))))).....((((.(((..(((((....-)))))...))).)))-)................. ( -25.20) >DroSec_CAF1 163934 95 + 1 AAAUGAACUUCCGCUCGGGCAAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU-UUCGCCACAUUCGCU-GCUCCCCCUCCACGAUCC ..(((((((((((((........)))..)))))))))).....((((.(((..(((((....-)))))...))).)))-)................. ( -25.20) >DroSim_CAF1 165820 95 + 1 AAAUGAACUUCCGCUCGGGCAAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU-UUCGCCACAUUCGCU-GCUCCCCCUCCACGAUCC ..(((((((((((((........)))..)))))))))).....((((.(((..(((((....-)))))...))).)))-)................. ( -25.20) >DroEre_CAF1 178122 95 + 1 AAAUGAACUUCCGCUCGGGCGAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU-UUCGCCACAUUCGCU-GCUCCCCCUCCGCGAUCC ..((((((((((......((....))..)))))))))).....((((.(((..(((((....-)))))...))).)))-)................. ( -25.40) >DroYak_CAF1 173178 92 + 1 AAAUGAACUUCCGCUCGGGCAAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU-UUCGCCACAUUCGCU-GCUCC---UCCACCAUCC ..(((((((((((((........)))..)))))))))).....((((.(((..(((((....-)))))...))).)))-)....---.......... ( -25.20) >DroAna_CAF1 170591 93 + 1 AAAUGAACUUCCGCCCGGGCAAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGAGAAAACUUUUCGCCACAUUCGCAGGCUCCCCCGCC----UCC ..(((((((((((((........)))..)))))))))).......((.(((.((.(((.....)))..)).))).))((((......)))----).. ( -26.10) >consensus AAAUGAACUUCCGCUCGGGCAAUGGCAAGGAAGUUCAUAAUUUCAGCUGAUUUGCGAAAACU_UUCGCCACAUUCGCU_GCUCCCCCUCCACGAUCC ..(((((((((((((........)))..)))))))))).......((.(((..(((((.....)))))...))).)).................... (-21.07 = -21.10 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:45:09 2006