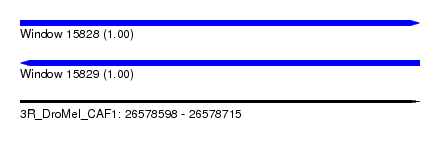
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,578,598 – 26,578,715 |
| Length | 117 |
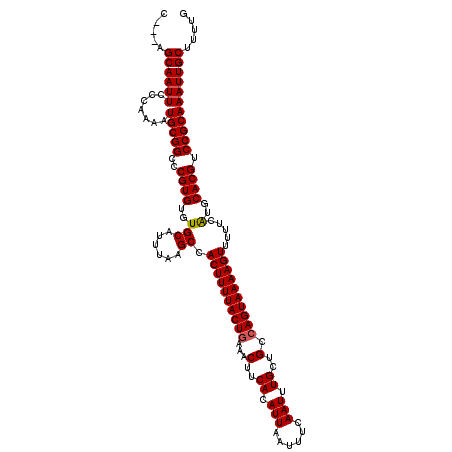
| Max. P | 0.999924 |

| Location | 26,578,598 – 26,578,715 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -31.50 |
| Energy contribution | -31.53 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.79 |
| SVM RNA-class probability | 0.999618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26578598 117 + 27905053 AAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACACGGGCCGCAUUUGGGGAAUUGCU---G ....((((((((.(.((.((((.......((((((((((((..((.((((....)))).))..))..))))))))))(((((............))))))))))).).))))))))---. ( -33.40) >DroPse_CAF1 208562 120 + 1 AAAAAGCAAUUUGCGGACGUGCACGGAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACACGGGCCGCAUUUGGGGAAUUGCUGCUG ....((((((((.(.((.((((.......((((((((((((..((.((((....)))).))..))..))))))))))(((((............))))))))))).).)))))))).... ( -33.40) >DroGri_CAF1 337418 117 + 1 CAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGGAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACACGGGCCGCAUUUUGGGAAUUGCA---G .....(((((((.((((.((((.......((((((((((((..((.((((....)))).))..))..))))))))))(((((............))))))))))))).))))))).---. ( -33.30) >DroWil_CAF1 193348 114 + 1 CAAAAGCAAUUUGCGGACGUGCAUGA-AAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACACGGGCCGCAUUUUGGGAAUUGC----- .....(((((((.((((.((((....-..((((((((((((..((.((((....)))).))..))..))))))))))(((((............))))))))))))).)))))))----- ( -31.90) >DroMoj_CAF1 216068 117 + 1 CAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGCUGAAGUAAAAGUGGCUUAAAUGCACACACGGGCCGCAUUUUGGGAAUUGCA---C .....(((((((.((((.((((.......(((((((((..((((..((((....)))).....)))).)))))))))(((((............))))))))))))).))))))).---. ( -35.40) >DroPer_CAF1 206540 120 + 1 AAAAAGCAAUUUGCGGACGUGCACGGAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACACGGGCCGCAUUUGGGGAAUUGCUGCUG ....((((((((.(.((.((((.......((((((((((((..((.((((....)))).))..))..))))))))))(((((............))))))))))).).)))))))).... ( -33.40) >consensus AAAAAGCAAUUUGCGGACGUGCAUGAAAAACUUUUACUGGCAGCAAAUUGAAAUUAAUGUGAAGUUUCAGUAAAAGUGGCUUAAAUGCACACACGGGCCGCAUUUGGGGAAUUGCU___G .....(((((((((((.((((........((((((((((((..((.((((....)))).))..))..)))))))))).((......))...))))..))))).......))))))..... (-31.50 = -31.53 + 0.03)

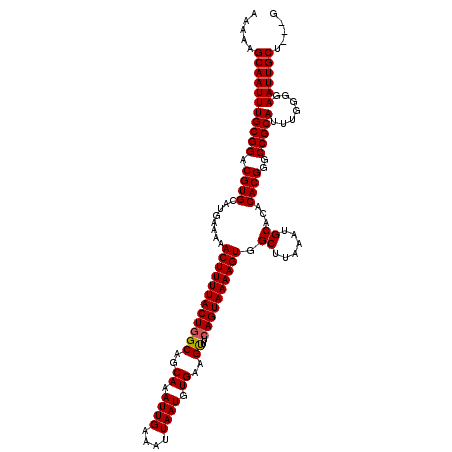

| Location | 26,578,598 – 26,578,715 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.22 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -27.97 |
| Energy contribution | -27.91 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.59 |
| SVM RNA-class probability | 0.999924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26578598 117 - 27905053 C---AGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUU .---(((((((.......(((((..(((((((((......)).((((((((((...(..((.(((......))).))..).)))))))))).....))))))).)))))))))))).... ( -29.21) >DroPse_CAF1 208562 120 - 1 CAGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUCCGUGCACGUCCGCAAAUUGCUUUUU ....(((((((.......(((((..((((..(((......)).((((((((((...(..((.(((......))).))..).)))))))))).....)..)))).)))))))))))).... ( -29.11) >DroGri_CAF1 337418 117 - 1 C---UGCAAUUCCCAAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUCCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUG .---.((((((.......(((((..(((((((((......)).((((((((((...(..((.(((......))).))..).)))))))))).....))))))).)))))))))))..... ( -28.41) >DroWil_CAF1 193348 114 - 1 -----GCAAUUCCCAAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUU-UCAUGCACGUCCGCAAAUUGCUUUUG -----((((((.......(((((..(((((((((......)).((((((((((...(..((.(((......))).))..).))))))))))..-..))))))).)))))))))))..... ( -28.41) >DroMoj_CAF1 216068 117 - 1 G---UGCAAUUCCCAAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUUCAGCUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUG .---.((((((.......(((((..(((((((((......)).(((((((((.((((...................))))..))))))))).....))))))).)))))))))))..... ( -31.42) >DroPer_CAF1 206540 120 - 1 CAGCAGCAAUUCCCCAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUCCGUGCACGUCCGCAAAUUGCUUUUU ....(((((((.......(((((..((((..(((......)).((((((((((...(..((.(((......))).))..).)))))))))).....)..)))).)))))))))))).... ( -29.11) >consensus C___AGCAAUUCCCAAAAUGCGGCCCGUGUGUGCAUUUAAGCCACUUUUACUGAAACUUCACAUUAAUUUCAAUUUGCUGCCAGUAAAAGUUUUUCAUGCACGUCCGCAAAUUGCUUUUG .....((((((.......(((((..((((..(((......)).((((((((((...(..((.(((......))).))..).)))))))))).....)..)))).)))))))))))..... (-27.97 = -27.91 + -0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:45:06 2006