
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,554,143 – 26,554,243 |
| Length | 100 |
| Max. P | 0.986081 |

| Location | 26,554,143 – 26,554,243 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -33.54 |
| Consensus MFE | -28.60 |
| Energy contribution | -30.22 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
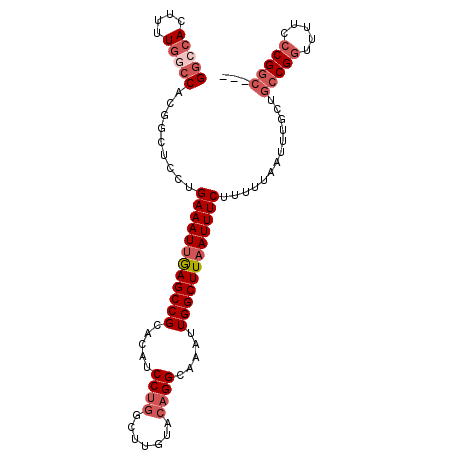
>3R_DroMel_CAF1 26554143 100 + 27905053 GGCCACUUUUGGCCACGGCUCCUGAAAUUGAGCCGCACAUCCUGGCUUGUACAGGCAAAUUGGCUUAAUUUCUUUUUAAUUUGCUGCCGGUUUUCCCGGC--- (((((....)))))..(((....((((((((((((.....((((.......)))).....))))))))))))..........)))(((((.....)))))--- ( -37.24) >DroVir_CAF1 178755 79 + 1 -----CUU--GGA-------GCUGAAAUGGAGCCGC--AUCC--GG-UGCCCAGGCAAAUUGGCUUAAUUUCUUUUUAAUUUGCUUCCGCUGCUUACG----- -----...--(.(-------((.(((.(((.((((.--...)--))-)..))).(((((((((............)))))))))))).))).).....----- ( -19.80) >DroSec_CAF1 138284 100 + 1 GGCCACUUUUGGCCACGGCUCCUGAAAUUAAGCCGCACAUCCUGGCUUGUACAGGCAAAUUGGCUUAAUUUCUUUUUAAUUUGCUGCCGGUUUUCCCGGC--- (((((....)))))..(((....((((((((((((.....((((.......)))).....))))))))))))..........)))(((((.....)))))--- ( -37.14) >DroSim_CAF1 139959 100 + 1 GGCCACUUUUGACCACGGCUCCUGAAAUUGAGCCGCACAUCCUGGCUUGUACAGGCAAAUUGGCUUAAUUUCUUUUUAAUUUGCUGCCGGUUUUCCCGGC--- ((((............))))...((((((((((((.....((((.......)))).....)))))))))))).............(((((.....)))))--- ( -31.70) >DroEre_CAF1 149377 100 + 1 GGCCACUUUUGGCCACGGCUCCUGAAAUUGAGCCGCACAUCCUGGCCUGUACAGGCAAAUUGGCUUAAUUUCUUUUUAAUUUGCUGCCGGUUUUCCCGGC--- (((((....)))))..(((....((((((((((((.....((((.......)))).....))))))))))))..........)))(((((.....)))))--- ( -37.24) >DroYak_CAF1 146071 103 + 1 GGCCACUUUUGGCCACCGCUCCUGAAAUUGAGCCGCACAUCCUGGCUUGUACAGGCAAAUUGGCUUAAUUUCUUUUUAAUUUGCUGCCGGUUUUCCCGGCUUU (((((....))))).........((((((((((((.....((((.......)))).....)))))))))))).............(((((.....)))))... ( -38.10) >consensus GGCCACUUUUGGCCACGGCUCCUGAAAUUGAGCCGCACAUCCUGGCUUGUACAGGCAAAUUGGCUUAAUUUCUUUUUAAUUUGCUGCCGGUUUUCCCGGC___ (((((....))))).........((((((((((((.....((((.......)))).....)))))))))))).............(((((.....)))))... (-28.60 = -30.22 + 1.61)


| Location | 26,554,143 – 26,554,243 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 86.93 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -27.35 |
| Energy contribution | -28.38 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.26 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
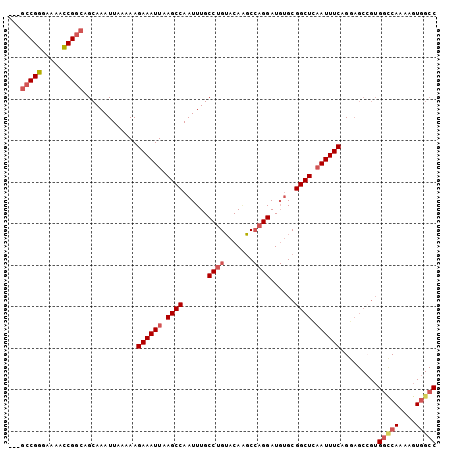
>3R_DroMel_CAF1 26554143 100 - 27905053 ---GCCGGGAAAACCGGCAGCAAAUUAAAAAGAAAUUAAGCCAAUUUGCCUGUACAAGCCAGGAUGUGCGGCUCAAUUUCAGGAGCCGUGGCCAAAAGUGGCC ---(((((.....))))).(((((((...((....)).....)))))))..(((((........)))))(((((........)))))..(((((....))))) ( -34.90) >DroVir_CAF1 178755 79 - 1 -----CGUAAGCAGCGGAAGCAAAUUAAAAAGAAAUUAAGCCAAUUUGCCUGGGCA-CC--GGAU--GCGGCUCCAUUUCAGC-------UCC--AAG----- -----....(((.((....))..........(((((..((((.((((((....)))-..--.)))--..))))..))))).))-------)..--...----- ( -20.90) >DroSec_CAF1 138284 100 - 1 ---GCCGGGAAAACCGGCAGCAAAUUAAAAAGAAAUUAAGCCAAUUUGCCUGUACAAGCCAGGAUGUGCGGCUUAAUUUCAGGAGCCGUGGCCAAAAGUGGCC ---(((((.....))))).............(((((((((((.((...((((.......))))..))..))))))))))).........(((((....))))) ( -38.20) >DroSim_CAF1 139959 100 - 1 ---GCCGGGAAAACCGGCAGCAAAUUAAAAAGAAAUUAAGCCAAUUUGCCUGUACAAGCCAGGAUGUGCGGCUCAAUUUCAGGAGCCGUGGUCAAAAGUGGCC ---(((((.....))))).....................((((.((((((.(((((........)))))(((((........)))))..)).))))..)))). ( -34.20) >DroEre_CAF1 149377 100 - 1 ---GCCGGGAAAACCGGCAGCAAAUUAAAAAGAAAUUAAGCCAAUUUGCCUGUACAGGCCAGGAUGUGCGGCUCAAUUUCAGGAGCCGUGGCCAAAAGUGGCC ---(((((.....))))).....................((((.(((((((....))))..((...((((((((........)))))))).)).))).)))). ( -36.40) >DroYak_CAF1 146071 103 - 1 AAAGCCGGGAAAACCGGCAGCAAAUUAAAAAGAAAUUAAGCCAAUUUGCCUGUACAAGCCAGGAUGUGCGGCUCAAUUUCAGGAGCGGUGGCCAAAAGUGGCC ...(((((.....))))).............((((((.((((.((...((((.......))))..))..)))).)))))).........(((((....))))) ( -34.50) >consensus ___GCCGGGAAAACCGGCAGCAAAUUAAAAAGAAAUUAAGCCAAUUUGCCUGUACAAGCCAGGAUGUGCGGCUCAAUUUCAGGAGCCGUGGCCAAAAGUGGCC ...(((((.....))))).............((((((.((((......((((.......))))......)))).)))))).........(((((....))))) (-27.35 = -28.38 + 1.03)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:45:00 2006