
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,549,065 – 26,549,156 |
| Length | 91 |
| Max. P | 0.877663 |

| Location | 26,549,065 – 26,549,156 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -17.92 |
| Consensus MFE | -12.84 |
| Energy contribution | -12.80 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26549065 91 + 27905053 CGCUCCAUUCUAGACUCACACUUUGUGUGGGCUUUGUUCUUGGCCAUAACGUCGCCAAUAUGAAGUACCCGCGAACAUUUCUUUUUCCUUA (((...........(((((((...)))))))(((..(..(((((.........))))).)..))).....))).................. ( -18.50) >DroSec_CAF1 133191 74 + 1 CGCUCCAUUCUAGACCCACACUUUGUGUGGGCUUUGUUCUUGGCCAUAAUGUCGCCAAUA-----------------UUCCUUUUUCCUUA ...........((.(((((((...)))))))))......(((((.........)))))..-----------------.............. ( -15.40) >DroSim_CAF1 134859 74 + 1 CGCUCCAUUCUAGACCCACACUUUGUGUGGGCUUUGUUCUUGGCCAUAAUGUCGCCAAUA-----------------UUCCUUUUUCCUUA ...........((.(((((((...)))))))))......(((((.........)))))..-----------------.............. ( -15.40) >DroEre_CAF1 144024 91 + 1 CGCUUUAUUCUAGACGCACACUUUGUGUGGGCUUUGUUCAUGGCCAUAACGUCGCCAAUUUUGAGUACCCGCGAACAUUUCUUUUUCCUUA (((..(((((..(((((((((...)))))((((........))))....)))).........)))))...))).................. ( -19.20) >DroYak_CAF1 140663 82 + 1 AGCUUUAUUCUAAACCCACACUUUGUGUGGGCUUUGUUCUUGGUCAUAACGUCGCCAAUUUGGAGUAU---------UUGUUGUUUCCUUA (((..((((((((((((((((...)))))))........(((((.........)))))))))))))).---------..)))......... ( -21.10) >consensus CGCUCCAUUCUAGACCCACACUUUGUGUGGGCUUUGUUCUUGGCCAUAACGUCGCCAAUAU__AGUA__________UUCCUUUUUCCUUA ..........(((((((((((...))))))).))))...(((((.........)))))................................. (-12.84 = -12.80 + -0.04)

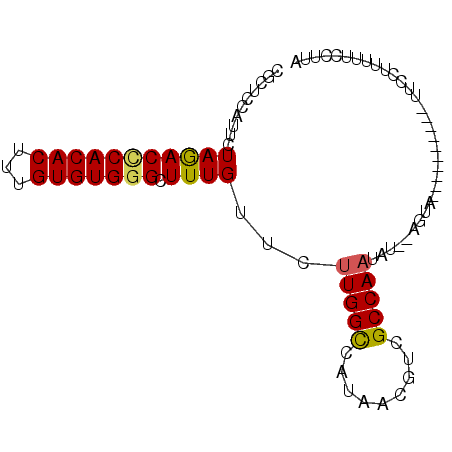

| Location | 26,549,065 – 26,549,156 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Mean single sequence MFE | -18.27 |
| Consensus MFE | -12.72 |
| Energy contribution | -12.96 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.637665 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26549065 91 - 27905053 UAAGGAAAAAGAAAUGUUCGCGGGUACUUCAUAUUGGCGACGUUAUGGCCAAGAACAAAGCCCACACAAAGUGUGAGUCUAGAAUGGAGCG ...............((((..((((........(((((.(.....).))))).......)))).(((.....)))...........)))). ( -18.96) >DroSec_CAF1 133191 74 - 1 UAAGGAAAAAGGAA-----------------UAUUGGCGACAUUAUGGCCAAGAACAAAGCCCACACAAAGUGUGGGUCUAGAAUGGAGCG ..............-----------------.....((..((((.(((...........((((((((...))))))))))).))))..)). ( -18.80) >DroSim_CAF1 134859 74 - 1 UAAGGAAAAAGGAA-----------------UAUUGGCGACAUUAUGGCCAAGAACAAAGCCCACACAAAGUGUGGGUCUAGAAUGGAGCG ..............-----------------.....((..((((.(((...........((((((((...))))))))))).))))..)). ( -18.80) >DroEre_CAF1 144024 91 - 1 UAAGGAAAAAGAAAUGUUCGCGGGUACUCAAAAUUGGCGACGUUAUGGCCAUGAACAAAGCCCACACAAAGUGUGCGUCUAGAAUAAAGCG ..............(((((((.(((.......))).))((((....(((..........)))(((((...)))))))))..)))))..... ( -16.80) >DroYak_CAF1 140663 82 - 1 UAAGGAAACAACAA---------AUACUCCAAAUUGGCGACGUUAUGACCAAGAACAAAGCCCACACAAAGUGUGGGUUUAGAAUAAAGCU ...(....).....---------..........((((((......)).)))).....((((((((((...))))))))))........... ( -18.00) >consensus UAAGGAAAAAGAAA__________UACU__AUAUUGGCGACGUUAUGGCCAAGAACAAAGCCCACACAAAGUGUGGGUCUAGAAUGGAGCG ....................................((..((((.(((...........((((((((...))))))))))).))))..)). (-12.72 = -12.96 + 0.24)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:58 2006