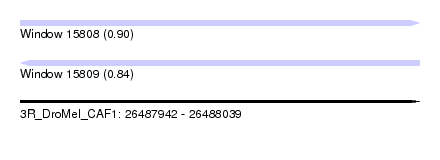
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,487,942 – 26,488,039 |
| Length | 97 |
| Max. P | 0.898383 |

| Location | 26,487,942 – 26,488,039 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -16.20 |
| Consensus MFE | -10.47 |
| Energy contribution | -10.97 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898383 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
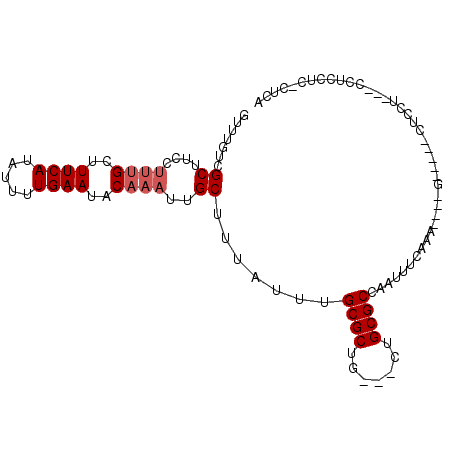
>3R_DroMel_CAF1 26487942 97 + 27905053 GUUUGUCGCUUCCUUUGCUUUCAUAUUUUGAAUACAAAUUGCUUUAUUUGCGCUG---CUGCGCCAAUUUGAAA----GUAGAGCUCCUUUCCUUCUUCUCUCA ............(((((((((((....(((.....((((......))))((((..---..)))))))..)))))----)))))).................... ( -18.60) >DroPse_CAF1 117863 87 + 1 GUUUGUCGCUUCCUUUGCUUUCAUAUUUUGAAUACAAAUUGCUUUAUUUGCGCUG---CUGCGCCAAUUUCAAA----A-----CUCU-----CUCCCCCCUCC ((((...((....((((..((((.....))))..))))..)).......((((..---..))))........))----)-----)...-----........... ( -13.40) >DroGri_CAF1 197711 93 + 1 --CAGUUGCUUCCCUGGCUUUCAUAUUUUGAAUACAAAUUGCUUUAUUUGCGCUG---CUGCGCCAAUUUGACAUACA--ACACAGCCUCAUCCCCCU----CA --.............((((((((.....))))..(((((((........((((..---..))))))))))).......--....))))..........----.. ( -17.00) >DroSim_CAF1 80121 97 + 1 GUUUGUCGCUUCCUUUGCUUUCAUAUUUUGAAUACAAAUUGCUUUAUUUGCGCUG---CUGCGCCAAUUUGAAA----GUAGAGCUCCUUUCCUUCUGCUCUCA ((((((.((.......)).((((.....)))).))))))(((((((((.((((..---..)))).)))...)))----)))((((............))))... ( -19.30) >DroAna_CAF1 80060 86 + 1 GUUUGUCGCUUCCU-UGCUUUCAUAUUUUGAAUACAAAUUGCUUUAUUUGCGCUGCUGCUGCGCCAAUUUCAAA----G-----CUCCC---CUUCUU-----A ((((((.((.....-.)).((((.....)))).)))))).((((((((.((((.......)))).)))...)))----)-----)....---......-----. ( -15.50) >DroPer_CAF1 115210 87 + 1 GUUUGUCGCUUCCUUUGCUUUCAUAUUUUGAAUACAAAUUGCUUUAUUUGCGCUG---CUGCGCCAAUUUCAAA----A-----CUCU-----CUCCCCCCUCC ((((...((....((((..((((.....))))..))))..)).......((((..---..))))........))----)-----)...-----........... ( -13.40) >consensus GUUUGUCGCUUCCUUUGCUUUCAUAUUUUGAAUACAAAUUGCUUUAUUUGCGCUG___CUGCGCCAAUUUCAAA____G_____CUCCU___CCUCCUC_CUCA .......((....((((..((((.....))))..))))..)).......((((.......))))........................................ (-10.47 = -10.97 + 0.50)

| Location | 26,487,942 – 26,488,039 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.34 |
| Mean single sequence MFE | -18.73 |
| Consensus MFE | -13.09 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.835294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
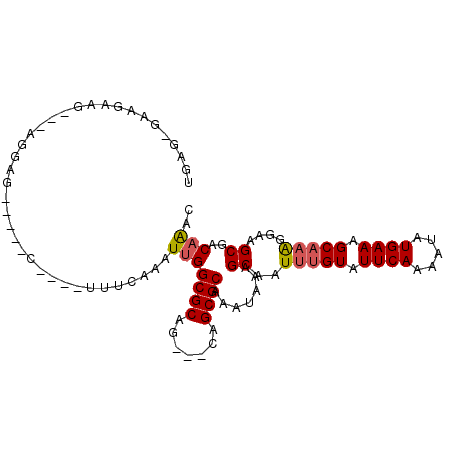
>3R_DroMel_CAF1 26487942 97 - 27905053 UGAGAGAAGAAGGAAAGGAGCUCUAC----UUUCAAAUUGGCGCAG---CAGCGCAAAUAAAGCAAUUUGUAUUCAAAAUAUGAAAGCAAAGGAAGCGACAAAC ((((((.(((.(........)))).)----)))))..(((((((..---..)))).......((..(((((.((((.....)))).)))))....))..))).. ( -21.40) >DroPse_CAF1 117863 87 - 1 GGAGGGGGGAG-----AGAG-----U----UUUGAAAUUGGCGCAG---CAGCGCAAAUAAAGCAAUUUGUAUUCAAAAUAUGAAAGCAAAGGAAGCGACAAAC ...........-----...(-----(----((((((....((((..---..))))..(((((....))))).))))))))......((.......))....... ( -15.40) >DroGri_CAF1 197711 93 - 1 UG----AGGGGGAUGAGGCUGUGU--UGUAUGUCAAAUUGGCGCAG---CAGCGCAAAUAAAGCAAUUUGUAUUCAAAAUAUGAAAGCCAGGGAAGCAACUG-- ..----..........((((((((--(((.((((.....))))..)---))))))..(((((....))))).((((.....)))))))).............-- ( -19.90) >DroSim_CAF1 80121 97 - 1 UGAGAGCAGAAGGAAAGGAGCUCUAC----UUUCAAAUUGGCGCAG---CAGCGCAAAUAAAGCAAUUUGUAUUCAAAAUAUGAAAGCAAAGGAAGCGACAAAC ..(((((............))))).(----((((......((((..---..))))...........(((((.((((.....)))).))))))))))........ ( -21.60) >DroAna_CAF1 80060 86 - 1 U-----AAGAAG---GGGAG-----C----UUUGAAAUUGGCGCAGCAGCAGCGCAAAUAAAGCAAUUUGUAUUCAAAAUAUGAAAGCA-AGGAAGCGACAAAC .-----......---.(..(-----(----(((.......((((.......))))...........(((((.((((.....)))).)))-)))))))..).... ( -18.70) >DroPer_CAF1 115210 87 - 1 GGAGGGGGGAG-----AGAG-----U----UUUGAAAUUGGCGCAG---CAGCGCAAAUAAAGCAAUUUGUAUUCAAAAUAUGAAAGCAAAGGAAGCGACAAAC ...........-----...(-----(----((((((....((((..---..))))..(((((....))))).))))))))......((.......))....... ( -15.40) >consensus UGAG_GAAGAAG___AGGAG_____C____UUUCAAAUUGGCGCAG___CAGCGCAAAUAAAGCAAUUUGUAUUCAAAAUAUGAAAGCAAAGGAAGCGACAAAC .....................................(((((((.......)))).......((..(((((.((((.....)))).)))))....))..))).. (-13.09 = -13.32 + 0.22)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:48 2006