| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,476,332 – 26,476,441 |
| Length | 109 |
| Max. P | 0.738444 |

| Location | 26,476,332 – 26,476,441 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 68.17 |
| Mean single sequence MFE | -26.58 |
| Consensus MFE | -15.22 |
| Energy contribution | -16.42 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.738444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26476332 109 + 27905053 AACUAAGCGCCUACGAUUGGUUUCGGGAGUUCUGCGAGAACGAUAUCAUUUACUUGUAGC--CGGAUUCUCGGAACAUUUAUUCAAUGCUUCA----AAUUUAUU--CAAAUUGCAG ....(((((..........(((((((((((((((((((..............))))))).--..))))))))))))..........)))))..----........--.......... ( -23.29) >DroSec_CAF1 68250 109 + 1 AACUUAGCGCCUACGAUUGGUUUCGGGAGUUCUGCGAGAACGAGAUCAUUUACUUGUAGC--CGGACUCCCGAAACUUUUAUUCAAUGAUUCA----AAUUUAUU--CAAAUUGCAG ......((......((..((((((((((((((.((....(((((........))))).))--.)))))))))))))).....))..(((....----.......)--))....)).. ( -32.30) >DroSim_CAF1 68699 108 + 1 AAUUAAGCGCCUACGAUUGGUUUCGGGAGUUUUGCGAGAACGAGAUCAUUUACUUGUAGC--CGGACUCUCGAAACUUUU-UUCAAUCAUUCA----AAUUUAUU--CAAAUUGCAG ......((......(((((((((((((((((..((....(((((........))))).))--..))))))))))))....-..))))).....----(((((...--.))))))).. ( -28.60) >DroEre_CAF1 76472 101 + 1 AGCUAAGCCGCUACAGUUGGUUUCGGGAGUUCUGCGAGAACGAGACCAUUUACUUGUAGA--CGGACUCUGGAAACUUUUCUAUAAUGAGCCCGUG--------------AUUUCUG ..........(((((((((((((((....(((...)))..))))))))...)).)))))(--(((.((((((((....)))))....))).)))).--------------....... ( -25.00) >DroYak_CAF1 72690 115 + 1 AACUAAGCGAAUACGAGUGGUUUCGCGAGUACUGCGAGAACGAGCCAAUUUACUUGUAGA--CGUACUCGCGAAACUUUUCUAUAAUGAGCCAGUGGAGUUUUUGCCAAAAUUGCAG ......((((....(((.((((((((((((((..(....(((((........))))).).--.)))))))))))))).))).............(((........)))...)))).. ( -34.40) >DroMoj_CAF1 94112 101 + 1 AGCUAGAGAAAUACCCAUGGUUUUACGAUUUCUGCUCAAACCAACGCAUUGAAAUCUAAACCUGC--UUCCGCACCU---A-CCGACUCUUUA----AAUUUAUG--UAUA----AG ...((((((.........(((((...(((((((((..........)))..)))))).)))))(((--....)))...---.-.....))))))----........--....----.. ( -15.90) >consensus AACUAAGCGCCUACGAUUGGUUUCGGGAGUUCUGCGAGAACGAGAUCAUUUACUUGUAGA__CGGACUCUCGAAACUUUUAUUCAAUGAUUCA____AAUUUAUU__CAAAUUGCAG ..................((((((((((((((((((((..............))))))).....)))))))))))))........................................ (-15.22 = -16.42 + 1.20)

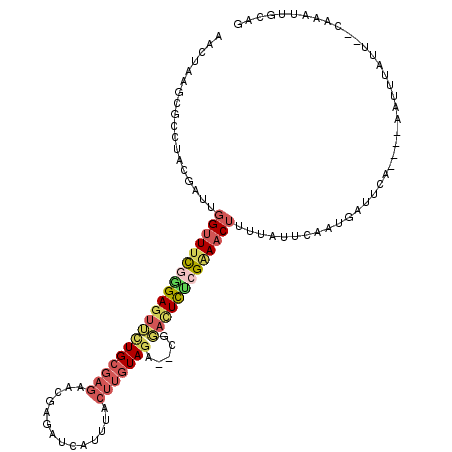

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:44 2006