| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,475,202 – 26,475,316 |
| Length | 114 |
| Max. P | 0.944879 |

| Location | 26,475,202 – 26,475,316 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -42.85 |
| Consensus MFE | -29.50 |
| Energy contribution | -29.31 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26475202 114 + 27905053 UCUCACCAUGUGGGACGUGGAGCAGCAGUUCUCUGUGUUCUUCGUGAGA---UCUAGCGAGCA---GGUCCGGGGCGAGCACAAAGAACUGCCUCCCGUUCACCCGAUCAUCCGGUACAA ........(((((((((.((((..((((((((.(((((((.((.((.((---(((.(....))---)))))).)).))))))).)))))))))))))))))..(((......))))))). ( -48.30) >DroVir_CAF1 89419 111 + 1 AUUAACCAUGUGGGAUGCGGAACGCGACUUUC------ACUUGAUGUAU---ACUGGCAAGGAGCCCGUGGAGGGUGUCUACGAGAAGGAUAACCCCGUUCAUCAGUUCGCUUUUUAUUU .........(((..(((((...))))..)..)------))((((((..(---((.(((.....))).)))(.((((((((.......)))).)))))...)))))).............. ( -25.00) >DroSec_CAF1 67170 114 + 1 UCUCACCAUGUGGGACGUGGAGCAGCAGUUCUCUGUGUUCUUCGUGAGA---UCUAGCGAGCA---GGUCCAGGGCGAGCACAAAGAACUGCCUCCCGUUCACCCGAUCAUCCGCUACAA ........(((((((((.((((..((((((((.(((((((.((.((.((---(((.(....))---)))))).)).))))))).))))))))))))))))....((......))))))). ( -45.40) >DroSim_CAF1 67623 114 + 1 UCUCACCAUGUGGGACGUGGAGCAGCAGUUCUCUGUGUUCUUCGUGAGA---UCUAGCGAGCA---GGUCCAGGGCGAGCACAAAGAACUUCCUCCCGUUCACCCGAUAAUCCGCUACAA ........(((((((((.((((....((((((.(((((((.((.((.((---(((.(....))---)))))).)).))))))).))))))..))))))))....((......))))))). ( -39.00) >DroEre_CAF1 75339 114 + 1 GCUCACCAUGUGGGACGUGGAGCGGCAGUUCUCUGUGUUCUUCGUGAGA---CCCAGCGAGCU---GGCCCAGGGUGAGCACAAAGAACUGCCGCCCGUUCACCCGACCUUCCGGUACAA ........((((((...(((.(((((((((((.(((((((.((.((.(.---.((((....))---)).))).)).))))))).))))))))))))))....))).(((....)))))). ( -51.30) >DroYak_CAF1 71566 117 + 1 GCUCACCAUGUGGGACGUGGAGCGGCAGUUCUCUGUGUUCUUCGUGCGACCACCCAGCGAGCA---GGCCCAGGAUGAGCACAGGGAACUGCCUCCCGUUCACCCGACCUUCCGCUACAA ((.(((...)))(((((.(((..(((((((((((((((((.((.((.(.((............---)).))).)).)))))))))))))))))))))))))............))..... ( -48.10) >consensus UCUCACCAUGUGGGACGUGGAGCAGCAGUUCUCUGUGUUCUUCGUGAGA___UCUAGCGAGCA___GGUCCAGGGCGAGCACAAAGAACUGCCUCCCGUUCACCCGAUCAUCCGCUACAA ........(((((((((.((((..((((((((.(((((((.((.((........................)).)).))))))).)))))))))))))))))...((......)).)))). (-29.50 = -29.31 + -0.19)

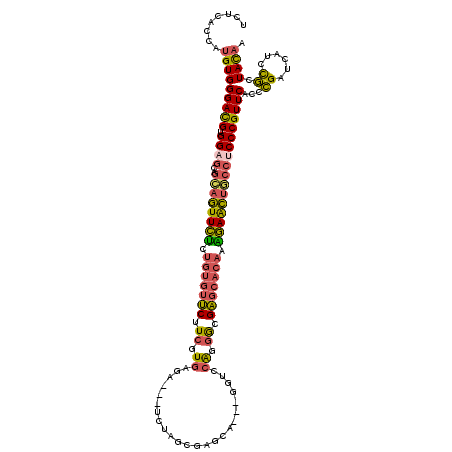

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:43 2006