
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,466,946 – 26,467,042 |
| Length | 96 |
| Max. P | 0.847702 |

| Location | 26,466,946 – 26,467,042 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -22.88 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.847702 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
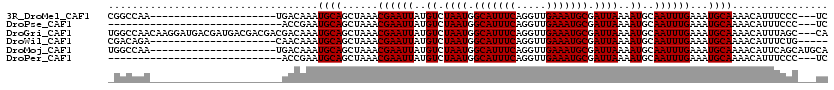
>3R_DroMel_CAF1 26466946 96 + 27905053 GA---GGGAAAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCA---------------------UUGGCCG ..---((.((((((...((....(.(((((...((((((.(((((((.....))))))).))))))..))))).)....)).)))))).)).---------------------....... ( -21.30) >DroPse_CAF1 95378 88 + 1 GA---GGGAAAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUCGGU----------------------------- ((---(.((((((.....))))))....((((........(((((((.....)))))))..((((((.......)))))).)))))))...----------------------------- ( -19.90) >DroGri_CAF1 154330 117 + 1 UG---GCUAAAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCGUCGUCGUCAUCGUCAUCCUUGUUGGCCA .(---((((((((...((((........)))).((((((.(((((((.....))))))).)))))).......))))))))).....((.((....)).))..((((.......)))).. ( -28.50) >DroWil_CAF1 66750 94 + 1 -----CAGAAAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUUG---------------------UCUGUCG -----..((((((.....))))))....((((........(((((((.....)))))))..((((((.......)))))).)))).......---------------------....... ( -19.40) >DroMoj_CAF1 83550 99 + 1 UGCAUGCUGAAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUCA---------------------UUGGCCA ((((.((((((((...((((........)))).((((((.(((((((.....))))))).)))))).......)))))))))))).......---------------------....... ( -28.30) >DroPer_CAF1 91770 88 + 1 GA---GGGAAAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUCGGU----------------------------- ((---(.((((((.....))))))....((((........(((((((.....)))))))..((((((.......)))))).)))))))...----------------------------- ( -19.90) >consensus GA___GGGAAAUGUUUUGCAUUUCAAAUUGCAUUUUAAUCGCAUUUCAACCUGAAAUGCCAUUAGACAUAAUUCGUUUAGCUGCAUUUGUC______________________UUGGCC_ .......((((((.....))))))....((((........(((((((.....)))))))..((((((.......)))))).))))................................... (-17.57 = -17.77 + 0.19)


| Location | 26,466,946 – 26,467,042 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.15 |
| Mean single sequence MFE | -20.34 |
| Consensus MFE | -18.00 |
| Energy contribution | -18.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.812591 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26466946 96 - 27905053 CGGCCAA---------------------UGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUUUCCC---UC .((..((---------------------((.....((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))...))))..))---.. ( -20.40) >DroPse_CAF1 95378 88 - 1 -----------------------------ACCGAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUUUCCC---UC -----------------------------......((((.......((......))((((.(((((((.....))))))).))))...))))....((((((.....))))))..---.. ( -18.40) >DroGri_CAF1 154330 117 - 1 UGGCCAACAAGGAUGACGAUGACGACGACGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUUUAGC---CA .(((.........((.((.((....)).)).))..((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...)))).........))---). ( -22.90) >DroWil_CAF1 66750 94 - 1 CGACAGA---------------------CAACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUUUCUG----- ...((((---------------------.......((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...)))).......))))----- ( -20.24) >DroMoj_CAF1 83550 99 - 1 UGGCCAA---------------------UGACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUUCAGCAUGCA .......---------------------.......(((((((..............((((.(((((((.....))))))).))))...((((........))))........))).)))) ( -21.70) >DroPer_CAF1 91770 88 - 1 -----------------------------ACCGAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUUUCCC---UC -----------------------------......((((.......((......))((((.(((((((.....))))))).))))...))))....((((((.....))))))..---.. ( -18.40) >consensus _GGCCAA______________________AACAAAUGCAGCUAAACGAAUUAUGUCUAAUGGCAUUUCAGGUUGAAAUGCGAUUAAAAUGCAAUUUGAAAUGCAAAACAUUUCCC___UC ...................................((((......((((((..((.((((.(((((((.....))))))).))))..))..))))))...))))................ (-18.00 = -18.00 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:40 2006