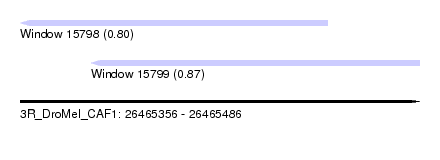
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,465,356 – 26,465,486 |
| Length | 130 |
| Max. P | 0.869961 |

| Location | 26,465,356 – 26,465,456 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 75.69 |
| Mean single sequence MFE | -17.01 |
| Consensus MFE | -12.37 |
| Energy contribution | -12.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.801497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
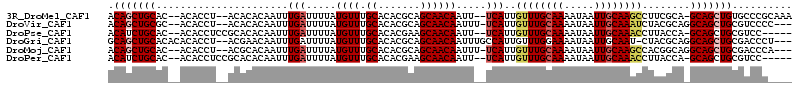
>3R_DroMel_CAF1 26465356 100 - 27905053 UUUAUGUUUGCACACGCAGCAACAAUU--UCAUUGUUUGCAAAAUAAUUGCAAGCCUUCGCA-GCAGCUGUGCCCGCAAAUCCAUCCACCUUC--CGUCCC---C-CUG .....((((((...((((((.......--.....((((((((.....)))))))).......-...))))))...))))))............--......---.-... ( -20.85) >DroVir_CAF1 74539 88 - 1 UUUAUGUUUGCACACGCAGCAACAAUUU-UCAUUGUUUGCAAAAUAAUUGCAAAUCUACGCAGGCAGCUGCGUCCCC-------------UGG--AAU--C---UGCUC ....((((.((....)))))).((..((-(((..((((((((.....))))))))..((((((....))))))....-------------)))--)).--.---))... ( -20.30) >DroPse_CAF1 93195 87 - 1 UUUAUGUUUGCACACGAAGCAACAAUU--UCAUUGUUUGCAAAAUAAUUGCAAACCUUACCA-GCAGCUGCGUCC-------------CGUUC--CGUUUC---C-CCG ...............(((((.......--.....((((((((.....)))))))).......-((....))....-------------.....--.)))))---.-... ( -14.40) >DroGri_CAF1 151851 92 - 1 UUUAUGUUUGCACACGCAGCAACAAUUUGCCAUUGUUUGGAAAAUAAUUGCAAU-CUACGCAGGCAGCUGCGACCCU-----------CCUUC--UAUCAC---UUCUC ........(((....)))((((..((((.(((.....))).))))..))))...-...(((((....))))).....-----------.....--......---..... ( -17.80) >DroAna_CAF1 58832 105 - 1 UUUAUGUUUGCACACGCAGCAACAAUU--UCAUUGUUUGCAAAAUAAUUGCAAACCUUCCCA-GCAGAUGUGCCCACAAAUCCUCUGUCAAUCCCCAACCCCAAC-CUA ....((((.((....))))))......--.....((((((((.....)))))))).......-((((((((....))).....))))).................-... ( -15.30) >DroPer_CAF1 89667 87 - 1 UUUAUGUUUGCACACGAAGCAACAAUU--UCAUUGUUUGCAAAAUAAUUGCAAACCUUACCA-GCAGCUGCGUCC-------------CGUUC--CGUCUC---C-CCG .............(((.(((.......--.....((((((((.....)))))))).......-((....))....-------------.))).--)))...---.-... ( -13.40) >consensus UUUAUGUUUGCACACGCAGCAACAAUU__UCAUUGUUUGCAAAAUAAUUGCAAACCUUCCCA_GCAGCUGCGUCC_C___________CCUUC__CAUCCC___C_CUG ..............((((((((((((.....))))))(((((.....)))))..............))))))..................................... (-12.37 = -12.82 + 0.44)

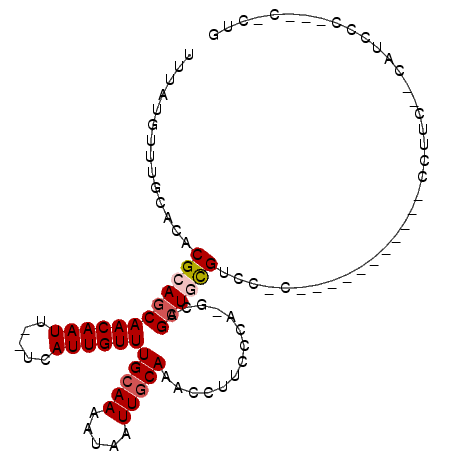
| Location | 26,465,379 – 26,465,486 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -20.23 |
| Energy contribution | -20.95 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.869961 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26465379 107 - 27905053 ACAGCUGCAC--ACACCU--ACACACAAUUUGAUUUUAUGUUUGCACACGCAGCAACAAUU--UCAUUGUUUGCAAAAUAAUUGCAAGCCUUCGCA-GCAGCUGUGCCCGCAAA ((((((((..--......--..........(((..((.((((.((....))))))..))..--)))..((((((((.....)))))))).......-))))))))......... ( -27.10) >DroVir_CAF1 74551 106 - 1 ACAGCUGCGC--ACACCU--ACACACAAUUUGAUUUUAUGUUUGCACACGCAGCAACAAUUU-UCAUUGUUUGCAAAAUAAUUGCAAAUCUACGCAGGCAGCUGCGUCCCC--- .(((((((((--......--..........(((.....(((((((....)))..))))....-)))..((((((((.....))))))))....))..))))))).......--- ( -24.30) >DroPse_CAF1 93210 104 - 1 ACAUCUGCAC--ACACCUCCGCACACAAUUUGAUUUUAUGUUUGCACACGAAGCAACAAUU--UCAUUGUUUGCAAAAUAAUUGCAAACCUUACCA-GCAGCUGCGUCC----- .((.((((..--........(((.(((...........))).)))....((((......))--))...((((((((.....)))))))).......-)))).)).....----- ( -18.80) >DroGri_CAF1 151867 108 - 1 GCAGCUGCACACACACCU--ACGAACAAUUUGAUUUUAUGUUUGCACACGCAGCAACAAUUUGCCAUUGUUUGGAAAAUAAUUGCAAU-CUACGCAGGCAGCUGCGACCCU--- ((((((((..........--.((((((((.............(((....)))((((....)))).))))))))........((((...-....))))))))))))......--- ( -30.60) >DroMoj_CAF1 82058 106 - 1 ACAGCUGCAC--ACACCU--ACGCACAAUUUGAUUUUAUGUUUGCACACGCAGCAACAAUUU-UCAUUGUUUGCAAAAUAAUUGCAAGCCACGGCAGGCAGCUGCGACCCA--- ...(.((((.--(((...--...((.....))......))).))))).((((((.(((((..-..)))))((((((.....))))))(((......))).)))))).....--- ( -26.80) >DroPer_CAF1 89682 104 - 1 ACAUCUGCAC--ACACCUCCGCACACAAUUUGAUUUUAUGUUUGCACACGAAGCAACAAUU--UCAUUGUUUGCAAAAUAAUUGCAAACCUUACCA-GCAGCUGCGUCC----- .((.((((..--........(((.(((...........))).)))....((((......))--))...((((((((.....)))))))).......-)))).)).....----- ( -18.80) >consensus ACAGCUGCAC__ACACCU__ACACACAAUUUGAUUUUAUGUUUGCACACGCAGCAACAAUU__UCAUUGUUUGCAAAAUAAUUGCAAACCUUCGCA_GCAGCUGCGUCCC____ .(((((((......................(((.....((((.((.......)))))).....)))..((((((((.....))))))))........))))))).......... (-20.23 = -20.95 + 0.72)
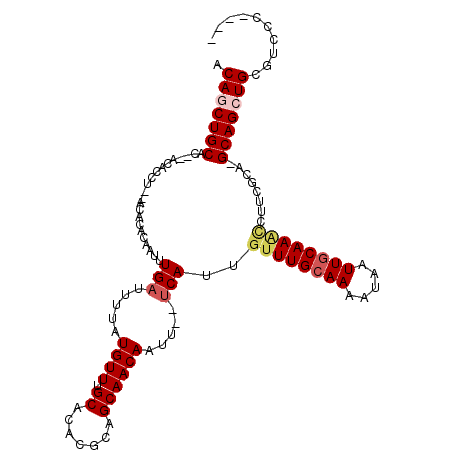
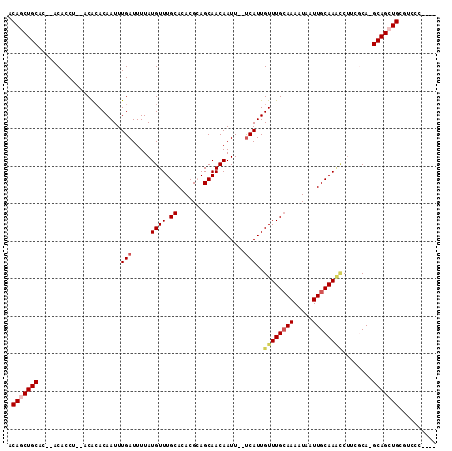
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:39 2006