
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
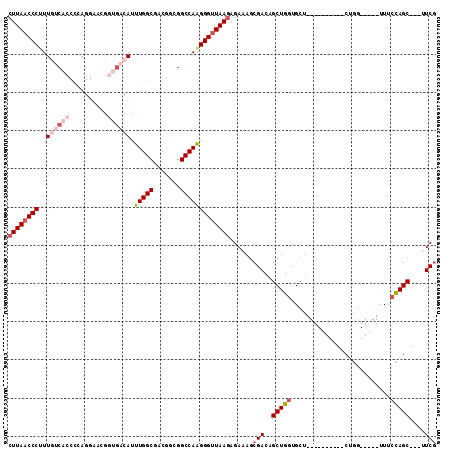
| Location | 26,458,383 – 26,458,477 |
| Length | 94 |
| Max. P | 0.601193 |

| Location | 26,458,383 – 26,458,477 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -21.01 |
| Energy contribution | -22.90 |
| Covariance contribution | 1.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.66 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
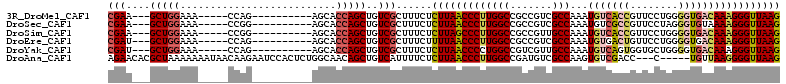
>3R_DroMel_CAF1 26458383 94 + 27905053 CUUAACCCUUUGUCACCCCAGGAACGGUGACAUUUGGCGACGGCGGCCAAGGGUUAAGAGAAAGCGACAGCUGGUGCU----------CUGG-----UUUCCAGC---UUCG ((((((((..(((((((........))))))).(((((.......)))))))))))))......(((.((((((.((.----------...)-----)..)))))---)))) ( -39.50) >DroSec_CAF1 50574 94 + 1 CUUAACCCUUUUACACCCUAGGAACGGCGACAUUUGGCGACGGCGGCCAAGGGUUAAGAGAAAGCGACAGCUGGUGCU----------CCGG-----UUUCCAGC---UUCG ((((((((.....(.((........)).)....(((((.......)))))))))))))......(((.((((((.((.----------...)-----)..)))))---)))) ( -30.20) >DroSim_CAF1 50799 94 + 1 CUUAACCCUUUGUCACCCCAGGAACGGUGACAUUUGGCAACGGCGGCCAAGGGCUAAGAGAAAGCGACAGCUGGUGCU----------CCGG-----UUUCCAGC---UUCG ((((.(((..(((((((........))))))).(((((.......)))))))).))))......(((.((((((.((.----------...)-----)..)))))---)))) ( -35.10) >DroEre_CAF1 58094 94 + 1 CUUAACCCUUUGUCACCCCAGGAACAGUCACAUUUGGCGACGGCGGCCAAGGGUUAAAAGAAAGCGACAGCUGGUGCU----------CUGG-----UUUCCAGC---AUCG .(((((((((.(((.(((((((..........)))))....)).))).))))))))).......(((..(((((.((.----------...)-----)..)))))---.))) ( -27.80) >DroYak_CAF1 52594 94 + 1 CUUAACCCUUUGUCACCCCAGCACCACUGACAUUUGGCAACGACGGCCAGGGGUUAAGAGAAAGCGACAGCUGGUGCU----------CUGG-----UUUCCAGC---AUCG ((((((((((.(((....(((.....)))....(((....))).))).))))))))))......(((..(((((.((.----------...)-----)..)))))---.))) ( -31.10) >DroAna_CAF1 51394 104 + 1 CUUAACCCCUUAACA-----G---GGUCGACACUUGGCGACAUCGGCCAAGGGUUAAGAGAAAAUGACAGCUGUUGCCAGAGUGGAUUCUUGUUAUUUUUUUAGCGUGUUCU ((((((((((....)-----)---((((((....((....))))))))..))))))))((((((((((((......((.....))....))))))))))))........... ( -28.60) >consensus CUUAACCCUUUGUCACCCCAGGAACGGUGACAUUUGGCGACGGCGGCCAAGGGUUAAGAGAAAGCGACAGCUGGUGCU__________CUGG_____UUUCCAGC___UUCG ((((((((..((((((..........)))))).(((((.......)))))))))))))......(((..(((((..........................)))))....))) (-21.01 = -22.90 + 1.89)


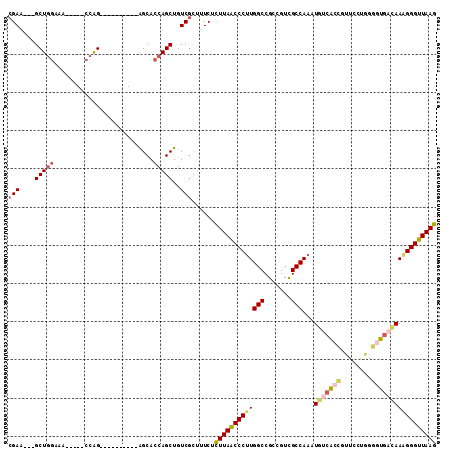
| Location | 26,458,383 – 26,458,477 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Mean single sequence MFE | -31.20 |
| Consensus MFE | -24.77 |
| Energy contribution | -25.72 |
| Covariance contribution | 0.95 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
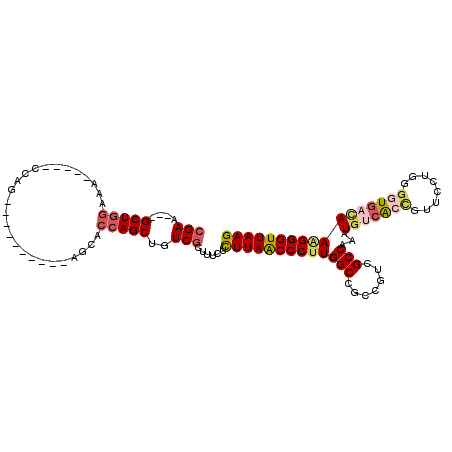
>3R_DroMel_CAF1 26458383 94 - 27905053 CGAA---GCUGGAAA-----CCAG----------AGCACCAGCUGUCGCUUUCUCUUAACCCUUGGCCGCCGUCGCCAAAUGUCACCGUUCCUGGGGUGACAAAGGGUUAAG ((((---(((((...-----....----------....)))))).)))......(((((((((((((.......)))...(((((((.(....).))))))))))))))))) ( -37.30) >DroSec_CAF1 50574 94 - 1 CGAA---GCUGGAAA-----CCGG----------AGCACCAGCUGUCGCUUUCUCUUAACCCUUGGCCGCCGUCGCCAAAUGUCGCCGUUCCUAGGGUGUAAAAGGGUUAAG ((((---(((((...-----....----------....)))))).)))......(((((((((((((.......)))......((((.(....).))))...)))))))))) ( -30.30) >DroSim_CAF1 50799 94 - 1 CGAA---GCUGGAAA-----CCGG----------AGCACCAGCUGUCGCUUUCUCUUAGCCCUUGGCCGCCGUUGCCAAAUGUCACCGUUCCUGGGGUGACAAAGGGUUAAG ((((---(((((...-----....----------....)))))).)))......(((((((((((((.......)))...(((((((.(....).))))))))))))))))) ( -37.20) >DroEre_CAF1 58094 94 - 1 CGAU---GCUGGAAA-----CCAG----------AGCACCAGCUGUCGCUUUCUUUUAACCCUUGGCCGCCGUCGCCAAAUGUGACUGUUCCUGGGGUGACAAAGGGUUAAG ((((---(((((...-----....----------....))))).)))).......(((((((((((..((.(((((.....))))).)).))...(....).))))))))). ( -29.70) >DroYak_CAF1 52594 94 - 1 CGAU---GCUGGAAA-----CCAG----------AGCACCAGCUGUCGCUUUCUCUUAACCCCUGGCCGUCGUUGCCAAAUGUCAGUGGUGCUGGGGUGACAAAGGGUUAAG ((((---(((((...-----....----------....))))).))))......((((((((.((((.......)))).((.((((.....)))).))......)))))))) ( -28.80) >DroAna_CAF1 51394 104 - 1 AGAACACGCUAAAAAAAUAACAAGAAUCCACUCUGGCAACAGCUGUCAUUUUCUCUUAACCCUUGGCCGAUGUCGCCAAGUGUCGACC---C-----UGUUAAGGGGUUAAG ......................((((...((.(((....)))..))....))))((((((.((((((.(....)))))))......((---(-----(....)))))))))) ( -23.90) >consensus CGAA___GCUGGAAA_____CCAG__________AGCACCAGCUGUCGCUUUCUCUUAACCCUUGGCCGCCGUCGCCAAAUGUCACCGUUCCUGGGGUGACAAAGGGUUAAG (((....(((((..........................)))))..)))......(((((((((((((.......)))...(((((((........))))))))))))))))) (-24.77 = -25.72 + 0.95)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:35 2006