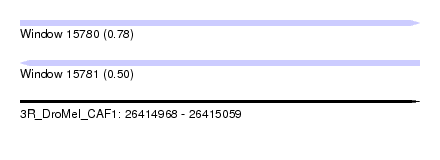
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,414,968 – 26,415,059 |
| Length | 91 |
| Max. P | 0.775053 |

| Location | 26,414,968 – 26,415,059 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -25.88 |
| Consensus MFE | -18.53 |
| Energy contribution | -20.03 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.775053 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26414968 91 + 27905053 UUGAUGAAAAUCAAAUAUCUUGGACGU---------GAAGGUCGAAGAUUGAAUGCCCUUCUCCUGGGAUUUCCCCAGGGGAAACCCUCUUUACCUUCUC (((((....))))).............---------((((((.(((((..(......)((((((((((.....))))))))))....))))))))))).. ( -30.30) >DroSec_CAF1 7211 91 + 1 UUGAUGAAAAUCAAAUAUCUUGGACGU---------GAAGGUCGAAGAUUGAAUGCCCUUCUCCUGGGAUUUCCACGCGGAAAACCUUCUUUACCUUCUC (((((....))))).............---------((((((.(((((..(....(((.......))).(((((....))))).)..))))))))))).. ( -24.10) >DroSim_CAF1 7065 91 + 1 UUGAUAAAAAUCAAAUAUCUUGGACGU---------GAAGGUCGAAGAUUGAAUGCCCUUCUCCUGGGAUUUCCACGGGGAAAACCUUCUUUACCUUCUC (((((....))))).............---------((((((.(((((..(....(((.......))).(((((....))))).)..))))))))))).. ( -21.60) >DroEre_CAF1 9963 91 + 1 UUGAUGAAAAUCAAGUGUCUUUGCCAC---------GAAGGCCGAAGAUGGAAUGCCCUUCUACUGGGGUUUCCACGGGGAAAACCUUCUUUACCUUCUC (((((....)))))(((.......)))---------(((((..((((((((((.(((((......)))))))))).(((.....)))))))).))))).. ( -30.00) >DroYak_CAF1 7290 91 + 1 UUGAUGAAAAUCAAAUAUCGUGGGUAU---------GAAGGCCGAAGGUUGAAUGCCCUUCUCCUGGGAUUUCCACGGGGAAAACCUUCUUCACCUUCUC (((((....))))).....(.((((..---------(((((..(((((........)))))(((((((....)).)))))....)))))...)))).).. ( -28.40) >DroAna_CAF1 7507 100 + 1 CUAAUGAAAAUCAGACAACUUUUACAGGGAUUAGAUCGAGGUCGAAGAUUGCUUGCCCUUCUCCGGGUAGUUCCAUAGGGAAAAGCAUCUCCACCUUCUC .....................................(((((.((.(((.((((((((......))))..((((....))))))))))))).)))))... ( -20.90) >consensus UUGAUGAAAAUCAAAUAUCUUGGACAU_________GAAGGUCGAAGAUUGAAUGCCCUUCUCCUGGGAUUUCCACGGGGAAAACCUUCUUUACCUUCUC (((((....)))))......................((((((.(((((.......(((.......))).(((((....)))))....))))))))))).. (-18.53 = -20.03 + 1.50)

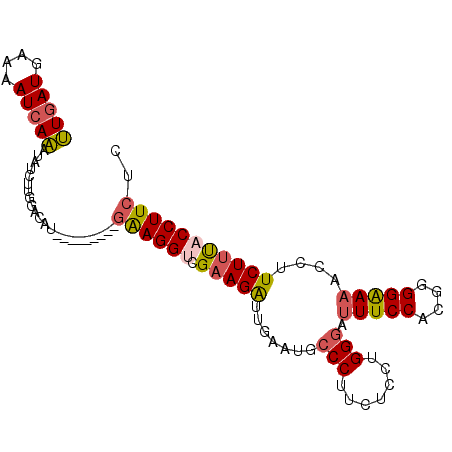

| Location | 26,414,968 – 26,415,059 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -17.07 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26414968 91 - 27905053 GAGAAGGUAAAGAGGGUUUCCCCUGGGGAAAUCCCAGGAGAAGGGCAUUCAAUCUUCGACCUUC---------ACGUCCAAGAUAUUUGAUUUUCAUCAA ..((((((...(((((((...((((((.....)))))).(((.....)))))))))).))))))---------.............(((((....))))) ( -28.30) >DroSec_CAF1 7211 91 - 1 GAGAAGGUAAAGAAGGUUUUCCGCGUGGAAAUCCCAGGAGAAGGGCAUUCAAUCUUCGACCUUC---------ACGUCCAAGAUAUUUGAUUUUCAUCAA ..((((((...(((((((.(((..(.((....))).)))(((.....)))))))))).))))))---------.............(((((....))))) ( -22.40) >DroSim_CAF1 7065 91 - 1 GAGAAGGUAAAGAAGGUUUUCCCCGUGGAAAUCCCAGGAGAAGGGCAUUCAAUCUUCGACCUUC---------ACGUCCAAGAUAUUUGAUUUUUAUCAA ..((((((...(((((((..((((.(((.....))).)....))).....))))))).))))))---------........((((.........)))).. ( -23.10) >DroEre_CAF1 9963 91 - 1 GAGAAGGUAAAGAAGGUUUUCCCCGUGGAAACCCCAGUAGAAGGGCAUUCCAUCUUCGGCCUUC---------GUGGCAAAGACACUUGAUUUUCAUCAA ((((((((...(((((((......((((((..(((.......)))..))))))....)))))))---------(((.......)))...)))))).)).. ( -25.00) >DroYak_CAF1 7290 91 - 1 GAGAAGGUGAAGAAGGUUUUCCCCGUGGAAAUCCCAGGAGAAGGGCAUUCAACCUUCGGCCUUC---------AUACCCACGAUAUUUGAUUUUCAUCAA .....((((((((.((....)).(((((.......(((.(((((........)))))..)))..---------....)))))........)))))))).. ( -26.22) >DroAna_CAF1 7507 100 - 1 GAGAAGGUGGAGAUGCUUUUCCCUAUGGAACUACCCGGAGAAGGGCAAGCAAUCUUCGACCUCGAUCUAAUCCCUGUAAAAGUUGUCUGAUUUUCAUUAG ((((((((((((((((((..((((.(((......)))....)))).)))).)))))).))))..(((..((..((.....))..))..)))))))..... ( -25.20) >consensus GAGAAGGUAAAGAAGGUUUUCCCCGUGGAAAUCCCAGGAGAAGGGCAUUCAAUCUUCGACCUUC_________ACGUCCAAGAUAUUUGAUUUUCAUCAA ..((((((...(((((((((((....))))..(((.......))).....))))))).))))))......................(((((....))))) (-17.07 = -17.38 + 0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:23 2006