| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,354,659 – 26,354,824 |
| Length | 165 |
| Max. P | 0.998888 |

| Location | 26,354,659 – 26,354,773 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.81 |
| Mean single sequence MFE | -40.99 |
| Consensus MFE | -36.07 |
| Energy contribution | -36.68 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
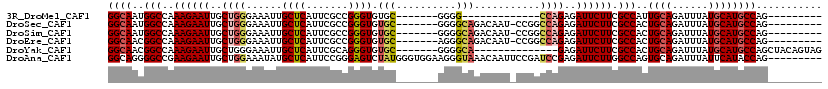
>3R_DroMel_CAF1 26354659 114 - 27905053 GCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCAUUGCCAAUGUUUGCCCUGGCAAGCAUGCUGCGGAAAAUGUUCGCUUAUCGGAAUCCAAGUGGGUCUCAACACUCACCGCA (((...(((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))................((((((......))))))))). ( -44.90) >DroSec_CAF1 76644 114 - 1 GCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCAUUGCCAAUGUUUGCCCUCGCAAGCAUGCUGCGGAAAAUGUUCGCUUAUCGGAAUCCAAGUGGGUCUCAACACUCACCGCA (((...(((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))................((((((......))))))))). ( -42.90) >DroSim_CAF1 81917 114 - 1 GCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCAUUGCCAAUGUUUGCCCUCGCAAGCAUGCUGCGGAAAAUGUUCGCUUAUCGGAAUCCAAGUGGGUCUCAACACUCACCGCA (((...(((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))................((((((......))))))))). ( -42.90) >DroEre_CAF1 84137 114 - 1 GCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCGUUGCCAAUGUUUGCCCUCGCAAGCAUGCUGCGGAAAAUGUUCGCUUAUCGGAAUCCAAGUGGGUCUCAGCCCUCACCGCA (((...(((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))(((....(..((((....))))..))))......))). ( -43.10) >DroYak_CAF1 80950 114 - 1 GCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCGUUGCCAAUGUUUGCCCUCGCAAGCACGCUGCGGAAAAUGUUCGCUUAUCGGAAUCCAAGUGGGUCUCAGCCCUCGCCGCA (((...(((((.(((((((((......(((((....)))))(((((((....))))))).)))).))))).)))))((.....((...))..(.((((....)))).)))))). ( -43.20) >DroAna_CAF1 96485 114 - 1 CGGAAUGAGCAUAUUUCCAGCAAUUCUUCGGCCCCUGCCAAUGUUUGCCCUCGGAAGUAUGCUGCGGAAAAUGUACGUUUAUCAGAAUUCAAGUGGGUCCCAGCACUUACCACA ..((((.(((((((((((.((((......(((....))).....))))....)))))))))))(((.........)))........))))..((((.............)))). ( -28.92) >consensus GCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCAUUGCCAAUGUUUGCCCUCGCAAGCAUGCUGCGGAAAAUGUUCGCUUAUCGGAAUCCAAGUGGGUCUCAACACUCACCGCA (((..((((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).))))))...............(((((........)))))))). (-36.07 = -36.68 + 0.61)


| Location | 26,354,694 – 26,354,793 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.72 |
| Mean single sequence MFE | -39.33 |
| Consensus MFE | -29.16 |
| Energy contribution | -29.94 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.30 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901149 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26354694 99 - 27905053 UCUCUGG-------------CCCC-------GCACACCCGGCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCAUUGCCAAUGUUUGCCCUGGCAAGCAUGCUGCGGAAAAUGUUCGCU .....((-------------(..(-------((.......)))...(((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))))) ( -39.20) >DroSec_CAF1 76679 111 - 1 UCUCUGGCCGG-AUUGUCUGCCCC-------GCACACCCGGCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCAUUGCCAAUGUUUGCCCUCGCAAGCAUGCUGCGGAAAAUGUUCGCU ......(((((-...((.(((...-------))).)))))))....(((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))... ( -44.00) >DroSim_CAF1 81952 111 - 1 UCUCUGGCCGG-AUUGUCUGCCCC-------GCACACCCGGCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCAUUGCCAAUGUUUGCCCUCGCAAGCAUGCUGCGGAAAAUGUUCGCU ......(((((-...((.(((...-------))).)))))))....(((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))... ( -44.00) >DroEre_CAF1 84172 111 - 1 UCUCUGGCCGG-AUUGUCUGCCCU-------GCACACCCGGCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCGUUGCCAAUGUUUGCCCUCGCAAGCAUGCUGCGGAAAAUGUUCGCU ......(((((-...((.(((...-------))).)))))))....(((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))... ( -44.00) >DroYak_CAF1 80985 98 - 1 UCUC--------------UGCCCC-------GCACACCCUGCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCGUUGCCAAUGUUUGCCCUCGCAAGCACGCUGCGGAAAAUGUUCGCU ....--------------.((..(-------(((.....))))...(((((.(((((((((......(((((....)))))(((((((....))))))).)))).))))).))))))). ( -37.00) >DroAna_CAF1 96520 119 - 1 UCUCGGAUCGGAAUUGUUUACCCUUCCACCCAUAGACUCCCGGAAUGAGCAUAUUUCCAGCAAUUCUUCGGCCCCUGCCAAUGUUUGCCCUCGGAAGUAUGCUGCGGAAAAUGUACGUU ....(((..((((....)).))..)))............(((.....(((((((((((.((((......(((....))).....))))....))))))))))).)))............ ( -27.80) >consensus UCUCUGGCCGG_AUUGUCUGCCCC_______GCACACCCGGCGAAUGAGCAAUUUCCCAGCAAUUCUUUGGCCAUUGCCAAUGUUUGCCCUCGCAAGCAUGCUGCGGAAAAUGUUCGCU ((.((((..............................)))).)).((((((.((((((((((.....(((((....)))))(((((((....)))))))))))).))))).)))))).. (-29.16 = -29.94 + 0.78)


| Location | 26,354,734 – 26,354,824 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -21.99 |
| Energy contribution | -22.72 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26354734 90 + 27905053 GGCAAUGGCCAAAGAAUUGCUGGGAAAUUGCUCAUUCGCCGGGUGUGC-------GGGG-------------CCAGAGAUUCUUCGCCAUUGCAGAUUUAUGCAUGCCAG--------- (((((((((..((((((..((((....((((.((((.....)))).))-------))..-------------))))..)))))).)))))((((......))))))))..--------- ( -35.20) >DroSec_CAF1 76719 102 + 1 GGCAAUGGCCAAAGAAUUGCUGGGAAAUUGCUCAUUCGCCGGGUGUGC-------GGGGCAGACAAU-CCGGCCAGAGAUUCUUCGCCACUGCAGAUUUAUGCAUGCCAG--------- ((((.((((..((((((..(((.(((........)))(((((((...(-------......)...))-))))))))..)))))).)))).((((......))))))))..--------- ( -37.40) >DroSim_CAF1 81992 102 + 1 GGCAAUGGCCAAAGAAUUGCUGGGAAAUUGCUCAUUCGCCGGGUGUGC-------GGGGCAGACAAU-CCGGCCAGAGAUUCUUCGCCACUGCAGAUUUAUGCAUGCCAG--------- ((((.((((..((((((..(((.(((........)))(((((((...(-------......)...))-))))))))..)))))).)))).((((......))))))))..--------- ( -37.40) >DroEre_CAF1 84212 102 + 1 GGCAACGGCCAAAGAAUUGCUGGGAAAUUGCUCAUUCGCCGGGUGUGC-------AGGGCAGACAAU-CCGGCCAGAGAUUCUUCGCCACUGCAGAUUUAUGCAUGCCAG--------- ((((..(((..((((((..(((.(((........)))(((((((.(((-------...))).))...-))))))))..)))))).)))..((((......))))))))..--------- ( -37.00) >DroYak_CAF1 81025 98 + 1 GGCAACGGCCAAAGAAUUGCUGGGAAAUUGCUCAUUCGCAGGGUGUGC-------GGGGCA--------------GAGAUUCUUCGCCACUGCAGAUUUAUGCAUGCCAGCUACAGUAG (((....)))........((((((((........)))(((((((.(((-------(((((.--------------.((...))..))).))))).)))).)))...)))))........ ( -32.00) >DroAna_CAF1 96560 110 + 1 GGCAGGGGCCGAAGAAUUGCUGGAAAUAUGCUCAUUCCGGGAGUCUAUGGGUGGAAGGGUAAACAAUUCCGAUCCGAGAUUCUUGGCCAGUGCAGAUUUAUUCAUACCAG--------- .(((..((((((.......((((((.........))))))((((((.(((((((((..(....)..)))).)))))))))))))))))..))).................--------- ( -37.50) >consensus GGCAAUGGCCAAAGAAUUGCUGGGAAAUUGCUCAUUCGCCGGGUGUGC_______GGGGCAGACAAU_CCGGCCAGAGAUUCUUCGCCACUGCAGAUUUAUGCAUGCCAG_________ ((((..(((..((((((..((((......((((.......)))).(((..........)))...........))))..)))))).)))..((((......))))))))........... (-21.99 = -22.72 + 0.72)
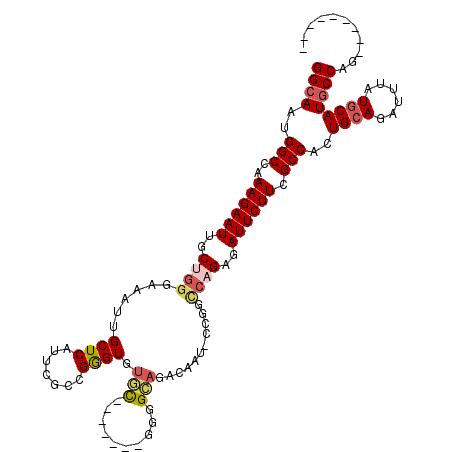
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:07 2006