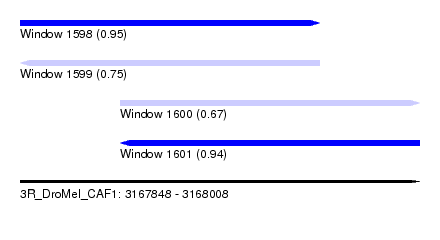
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,167,848 – 3,168,008 |
| Length | 160 |
| Max. P | 0.949254 |

| Location | 3,167,848 – 3,167,968 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -41.52 |
| Consensus MFE | -41.38 |
| Energy contribution | -41.38 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.95 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.949254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3167848 120 + 27905053 GUAGAAGUCAGCAAUUAUCUAAAAAGCGAGCAGCGGCGACAUCCUGCGAAAAGGAUAAUAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUU .((((.(........).))))....((.((((((((.(((((((((((((...........(((((....))))).)))))).)))((.....))....)))))))))))).))...... ( -41.20) >DroSec_CAF1 15927 120 + 1 GUAGAAGUCAGCAAUUAUCUAAAAAGCGAGCAGCGGCGACAUCCUGCGAAAAGGAUAACAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUU .((((.(........).))))....((.((((((((.(((((((((((((..(.....)..(((((....))))).)))))).)))((.....))....)))))))))))).))...... ( -42.40) >DroSim_CAF1 18691 120 + 1 GUAGAAGUCAGCAAUUAUCUAAAAAGCGAGCAGCGGCGACAUCCUGCGAAAAGGAUAAUAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUU .((((.(........).))))....((.((((((((.(((((((((((((...........(((((....))))).)))))).)))((.....))....)))))))))))).))...... ( -41.20) >DroEre_CAF1 7587 119 + 1 GUAGAAGUCAGCAAUUAUCUAAAAAGCGAGCAGCGGCGACAUCCUGCGAA-AGGAUAAUAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUU .((((.(........).))))....((.((((((((.(((((((((((((-..........(((((....))))).)))))).)))((.....))....)))))))))))).))...... ( -41.30) >consensus GUAGAAGUCAGCAAUUAUCUAAAAAGCGAGCAGCGGCGACAUCCUGCGAAAAGGAUAAUAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUU .((((.(........).))))....((.((((((((.(((((((((((((...........(((((....))))).)))))).)))((.....))....)))))))))))).))...... (-41.38 = -41.38 + 0.00)

| Location | 3,167,848 – 3,167,968 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.17 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -35.08 |
| Energy contribution | -35.08 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754575 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3167848 120 - 27905053 AAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUAUUAUCCUUUUCGCAGGAUGUCGCCGCUGCUCGCUUUUUAGAUAAUUGCUGACUUCUAC ......((.(((((((((((.((........))))))..((....(((((....)))))....((((((......))))))..))))))))).))....((((............)))). ( -35.10) >DroSec_CAF1 15927 120 - 1 AAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUGUUAUCCUUUUCGCAGGAUGUCGCCGCUGCUCGCUUUUUAGAUAAUUGCUGACUUCUAC ......((.(((((((((((.((........))))))..((....(((((....)))))..(.((((((......)))))).)))))))))).))....((((............)))). ( -35.40) >DroSim_CAF1 18691 120 - 1 AAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUAUUAUCCUUUUCGCAGGAUGUCGCCGCUGCUCGCUUUUUAGAUAAUUGCUGACUUCUAC ......((.(((((((((((.((........))))))..((....(((((....)))))....((((((......))))))..))))))))).))....((((............)))). ( -35.10) >DroEre_CAF1 7587 119 - 1 AAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUAUUAUCCU-UUCGCAGGAUGUCGCCGCUGCUCGCUUUUUAGAUAAUUGCUGACUUCUAC ......((.(((((((((((.((........))))))..((....(((((....)))))....((((((-.....))))))..))))))))).))....((((............)))). ( -34.90) >consensus AAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUAUUAUCCUUUUCGCAGGAUGUCGCCGCUGCUCGCUUUUUAGAUAAUUGCUGACUUCUAC ......((.(((((((((((.((........))))))..((....(((((....)))))....((((((......))))))..))))))))).))....((((............)))). (-35.08 = -35.08 + -0.00)

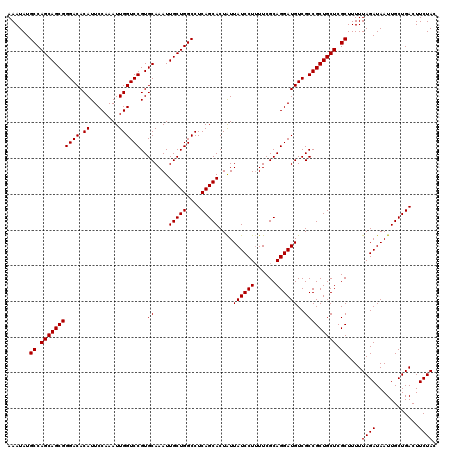
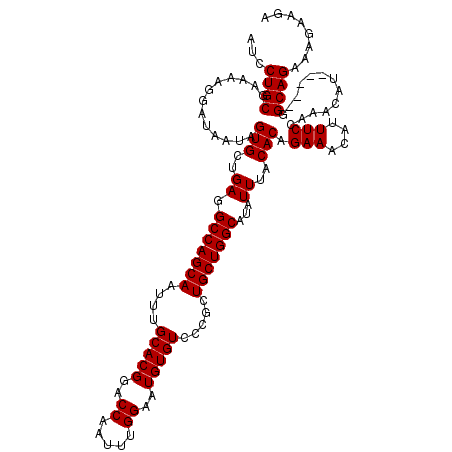
| Location | 3,167,888 – 3,168,008 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -30.75 |
| Consensus MFE | -29.30 |
| Energy contribution | -29.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.666323 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3167888 120 + 27905053 AUCCUGCGAAAAGGAUAAUAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUUACACAGAAACAUUUCCAAACAGAAAUAGGCAGAAAGAAGA ...((((.............(((..((.(((((((....(((((..((.....))..))))).....)))))))...))..)))......(((((......)))))..))))........ ( -31.70) >DroSec_CAF1 15967 115 + 1 AUCCUGCGAAAAGGAUAACAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUUACACAGAAACAUUUCCAAACAU-----GGCAGAAAGAAGA (((((......)))))....(((..((.(((((((....(((((..((.....))..))))).....)))))))...))..))).......((((((....)-----)).)))....... ( -30.50) >DroSim_CAF1 18731 115 + 1 AUCCUGCGAAAAGGAUAAUAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUUACACAGAAACAUUUCCAAACAU-----GGCAGAAAGAAGA (((((......)))))....(((..((.(((((((....(((((..((.....))..))))).....)))))))...))..))).......((((((....)-----)).)))....... ( -30.50) >DroEre_CAF1 7627 114 + 1 AUCCUGCGAA-AGGAUAAUAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUUACACAGAAACAUUUCCAAACAU-----GGCAGAAAGAAGA (((((.....-)))))....(((..((.(((((((....(((((..((.....))..))))).....)))))))...))..))).......((((((....)-----)).)))....... ( -30.30) >consensus AUCCUGCGAAAAGGAUAAUAGUGCUGAGGCCAGCAAUUUGCACGGACCAAUUUGGAAUGUGUCCCGCUGCUGGCAUAUUUACACAGAAACAUUUCCAAACAU_____GGCAGAAAGAAGA ...((((.............(((..((.(((((((....(((((..((.....))..))))).....)))))))...))..))).(((....))).............))))........ (-29.30 = -29.30 + 0.00)

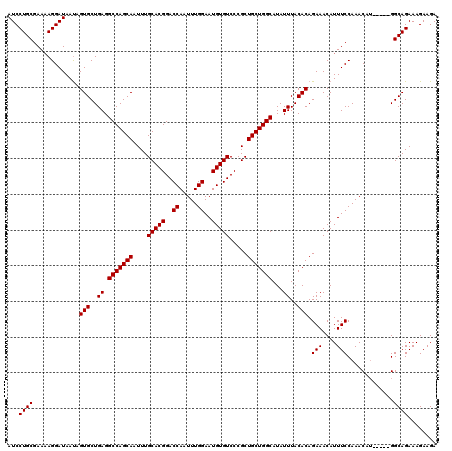
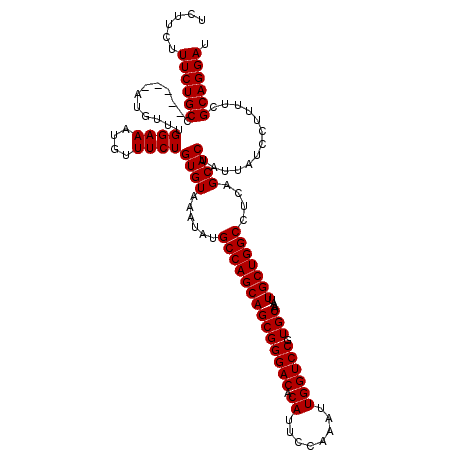
| Location | 3,167,888 – 3,168,008 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.60 |
| Mean single sequence MFE | -33.10 |
| Consensus MFE | -31.90 |
| Energy contribution | -31.90 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.942395 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3167888 120 - 27905053 UCUUCUUUCUGCCUAUUUCUGUUUGGAAAUGUUUCUGUGUAAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUAUUAUCCUUUUCGCAGGAU ......((((((.(((((((....))))))).....((((......((((((((((((((.((........)))))).)))....)))))))....)))).............)))))). ( -35.10) >DroSec_CAF1 15967 115 - 1 UCUUCUUUCUGCC-----AUGUUUGGAAAUGUUUCUGUGUAAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUGUUAUCCUUUUCGCAGGAU ......(((((((-----(....))((((.((..(.((((......((((((((((((((.((........)))))).)))....)))))))....)))).)..))...)))))))))). ( -32.60) >DroSim_CAF1 18731 115 - 1 UCUUCUUUCUGCC-----AUGUUUGGAAAUGUUUCUGUGUAAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUAUUAUCCUUUUCGCAGGAU ......((((((.-----......(((...((....((((......((((((((((((((.((........)))))).)))....)))))))....)))).))..))).....)))))). ( -32.20) >DroEre_CAF1 7627 114 - 1 UCUUCUUUCUGCC-----AUGUUUGGAAAUGUUUCUGUGUAAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUAUUAUCCU-UUCGCAGGAU ......(((((((-----(....))((((.(.....((((......((((((((((((((.((........)))))).)))....)))))))....))))......).)-))))))))). ( -32.50) >consensus UCUUCUUUCUGCC_____AUGUUUGGAAAUGUUUCUGUGUAAAUAUGCCAGCAGCGGGACACAUUCCAAAUUGGUCCGUGCAAAUUGCUGGCCUCAGCACUAUUAUCCUUUUCGCAGGAU ......((((((............((((....))))((((......((((((((((((((.((........)))))).)))....)))))))....)))).............)))))). (-31.90 = -31.90 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:43 2006