
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,351,903 – 26,352,001 |
| Length | 98 |
| Max. P | 0.660806 |

| Location | 26,351,903 – 26,352,001 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 75.83 |
| Mean single sequence MFE | -18.05 |
| Consensus MFE | -13.32 |
| Energy contribution | -13.93 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
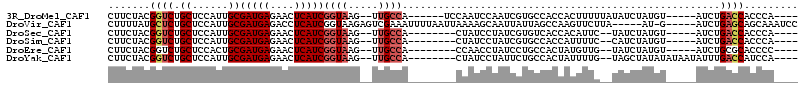
>3R_DroMel_CAF1 26351903 98 - 27905053 CUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAG--UUGCCA------UCCAAUCCAAUCGUGCCACCACUUUUUAUAUCUAUGU-----AUCUGACCACCCA---- .......((((.((......))(((((....)))))((((.(--(((...------.......))))..))))....................-----....)))).....---- ( -17.70) >DroVir_CAF1 74379 104 - 1 CUUUUAUGCUCUGCUCCAUUGCGAUGAGACCUCAUCGGUAAGAGUCGAAAUUUUAAUUAAAAGCAAUUAUUAGCCAAGUUCUUA-----AU-G-----AUCUGAGCAGCAAAUCC ......((((..((((.....((((((....)))))).((((((.(....(..(((((......)))))..).....)))))))-----..-.-----....))))))))..... ( -19.90) >DroSec_CAF1 73953 94 - 1 CUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAG--UUGCCA--------CUAUCCUAUCGUGUCACCACAUUC--UAUCUAUGU-----AUCUGACCACCCA---- .......((((.((......))(((((....)))))(((...--..))).--------..........(((....)))....--.........-----....)))).....---- ( -17.40) >DroSim_CAF1 79180 94 - 1 CUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAG--UUGCCA--------CUAUCCUAUCGUGCCACCAUUUUC--CAUCUAUGU-----AUCUGACCACCCA---- .......((((.((......))(((((....)))))((.(((--(((.((--------(.........))).))..)))).)--)........-----....)))).....---- ( -17.70) >DroEre_CAF1 81371 94 - 1 CUUCUACGGUCUGCUCCACUGCGAUGAGAACUCAUCGGUAAG--UUGCCA--------CCAACCUAUCCUGCCACUAUGUUG--UAUCUAUGU-----AUCUGCGCACCCC---- .......(((.(((.....((((((((....)))))((((.(--(((...--------.)))).))))..............--.......))-----)...))).)))..---- ( -17.10) >DroYak_CAF1 78143 99 - 1 CUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAG--UUGCCA--------CUAUCCUAUUCUGCCACUAUUUUG--UAGCUAUAUAUAAUAUUUGACCAUCCA---- .......((((.((......))(((((....)))))(((...--..))).--------.(((..(((...((.((......)--).))...)))..)))...)))).....---- ( -18.50) >consensus CUUCUACGGUCUGCUCCAUUGCGAUGAGAACUCAUCGGUAAG__UUGCCA________CUAUCCUAUCGUGCCACCAUUUUC__UAUCUAUGU_____AUCUGACCACCCA____ .......((((.((......))(((((....)))))((((.....)))).....................................................))))......... (-13.32 = -13.93 + 0.61)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:44:05 2006