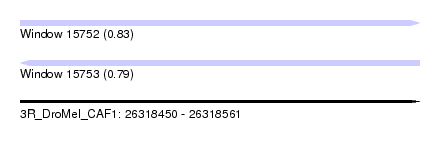
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,318,450 – 26,318,561 |
| Length | 111 |
| Max. P | 0.826151 |

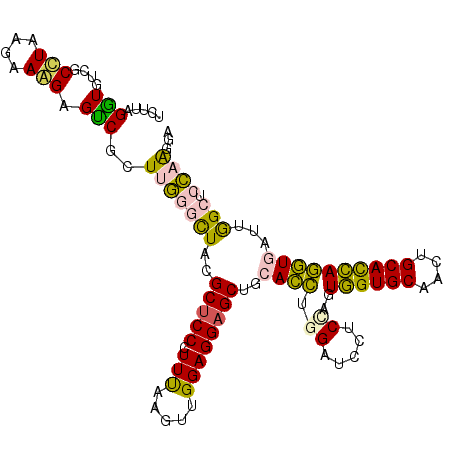
| Location | 26,318,450 – 26,318,561 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -21.57 |
| Energy contribution | -21.93 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26318450 111 + 27905053 UCUUUGGAGCCAAUAACCUGAUGCAAUUGCACCACCAGUGGGUCCAGGUGGAGCUCCUCCAACUUAAAUGGUGCGUAGCCCAAGCGACUCUUCCUUAAGCGACACCUAAGA .....((((((....(((((.(((....)))(((....)))...))))).).))))).....((((...(((((((.((....))((....)).....))).)))))))). ( -30.70) >DroPse_CAF1 37302 111 + 1 UCCCGCUGGCUGACCAUGUCGUGCACCUGCACAAUGGGAGGAUCUGCAUGCAGCUCCUCCAGCUUGAAGGGAGCGUAGUCGAAGCGCCUCCUUUGCAGGCGGCAGCGAAGA ...(((((.(....(((...((((....)))).)))((((((.(((....))).)))))).(((((((((((((((.......))).))))))).)))))).))))).... ( -50.70) >DroSim_CAF1 45188 111 + 1 UCAUUGGAGCCAAUAACCUGAUGCAGUUGCACCACUAGAGGGUCCAGGUGUAGCUCCUCCAACUUAAAAGGAGCGUAGCCCAAGCGACUCUUCCUUAAGCGACACCUAAGA .....(((.((..((...((.(((....))).)).))..)).)))((((((.((((((..........))))))...((..(((.((....)))))..)).)))))).... ( -31.00) >DroEre_CAF1 48325 111 + 1 UCUUUGGAGCCAAUCACCUGGUGCAGCUGCACCACUGGGGGAUCCAGGUGCAGCUCCUCCAACUUAAAGGGAGCGUAACCCAAGCGACUCCUCCAAAGGCGACACCUGAGA ((((.((.(((.......((((((....)))))).((((((((((.(((...((((((..........))))))...)))...).)).)))))))..)))....)).)))) ( -42.30) >DroYak_CAF1 44026 111 + 1 UCCUUGGCGCCAAUCACCUGAUGCAGUUGCACCAGCAGCGGAUCCAGGUGCAGCUCCUCCAACUUAAAAGGAGCGUAGCCCAAGCGACUCUUUCUAAGGCGACAUCUGAGA ..((..(((((...((((((((.(.(((((....)))))).)).))))))..((((((..........))))))((.((....)).)).........))))....)..)). ( -35.80) >DroPer_CAF1 37152 111 + 1 UCCCGCUGGCUGACCAUGUCGUGCACCUGCACAAUGGGAGGAUCUGCAUGCAGCUCCUCCAGCUUGAAGGGAGCGUAGUCGAAGCGCCUCCUUUGCAGGCGGCAGCGAAGA ...(((((.(....(((...((((....)))).)))((((((.(((....))).)))))).(((((((((((((((.......))).))))))).)))))).))))).... ( -50.70) >consensus UCCUUGGAGCCAAUCACCUGAUGCAGCUGCACCACCAGAGGAUCCAGGUGCAGCUCCUCCAACUUAAAGGGAGCGUAGCCCAAGCGACUCCUCCUAAGGCGACACCUAAGA .....(((((....((((((.(((....)))((......))...))))))..))))).....((((..((((((((.......))).)))))...))))............ (-21.57 = -21.93 + 0.37)

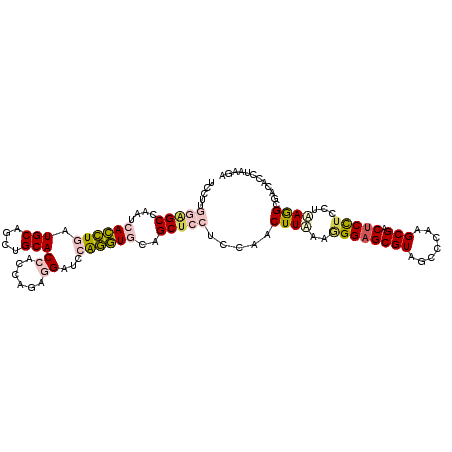
| Location | 26,318,450 – 26,318,561 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Mean single sequence MFE | -43.48 |
| Consensus MFE | -22.90 |
| Energy contribution | -21.13 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.788916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26318450 111 - 27905053 UCUUAGGUGUCGCUUAAGGAAGAGUCGCUUGGGCUACGCACCAUUUAAGUUGGAGGAGCUCCACCUGGACCCACUGGUGGUGCAAUUGCAUCAGGUUAUUGGCUCCAAAGA ((((.(((((.(((((((((....)).)))))))...)))))............((((((..((((((..(((....)))(((....)))))))))....)))))).)))) ( -38.20) >DroPse_CAF1 37302 111 - 1 UCUUCGCUGCCGCCUGCAAAGGAGGCGCUUCGACUACGCUCCCUUCAAGCUGGAGGAGCUGCAUGCAGAUCCUCCCAUUGUGCAGGUGCACGACAUGGUCAGCCAGCGGGA ..(((((((.((((((((((((..(((.........)))..))))(((...((((((.(((....))).))))))..)))))))))))...(((...)))...))))))). ( -46.40) >DroSim_CAF1 45188 111 - 1 UCUUAGGUGUCGCUUAAGGAAGAGUCGCUUGGGCUACGCUCCUUUUAAGUUGGAGGAGCUACACCUGGACCCUCUAGUGGUGCAACUGCAUCAGGUUAUUGGCUCCAAUGA ....((((((.(((((((((....)).)))))))...((((((((......)))))))).))))))(((.((..(((((((((....))))))..)))..)).)))..... ( -41.10) >DroEre_CAF1 48325 111 - 1 UCUCAGGUGUCGCCUUUGGAGGAGUCGCUUGGGUUACGCUCCCUUUAAGUUGGAGGAGCUGCACCUGGAUCCCCCAGUGGUGCAGCUGCACCAGGUGAUUGGCUCCAAAGA .............((((((((.((((((((((((....((((.........)))).((((((((((((.....)))..))))))))))).))))))))))..)))))))). ( -48.80) >DroYak_CAF1 44026 111 - 1 UCUCAGAUGUCGCCUUAGAAAGAGUCGCUUGGGCUACGCUCCUUUUAAGUUGGAGGAGCUGCACCUGGAUCCGCUGCUGGUGCAACUGCAUCAGGUGAUUGGCGCCAAGGA .......((.((((........((((((((((((...((((((((......))))))))((((((.((........))))))))...)).)))))))))))))).)).... ( -40.00) >DroPer_CAF1 37152 111 - 1 UCUUCGCUGCCGCCUGCAAAGGAGGCGCUUCGACUACGCUCCCUUCAAGCUGGAGGAGCUGCAUGCAGAUCCUCCCAUUGUGCAGGUGCACGACAUGGUCAGCCAGCGGGA ..(((((((.((((((((((((..(((.........)))..))))(((...((((((.(((....))).))))))..)))))))))))...(((...)))...))))))). ( -46.40) >consensus UCUUAGGUGUCGCCUAAGAAAGAGUCGCUUGGGCUACGCUCCCUUUAAGUUGGAGGAGCUGCACCUGGAUCCUCCAGUGGUGCAACUGCACCAGGUGAUUGGCUCCAAGGA .....(((....(((....))).)))..(((((((..(((((.(((.....))))))))..((((.((.....))..((((((....))))))))))...))).))))... (-22.90 = -21.13 + -1.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:43:58 2006