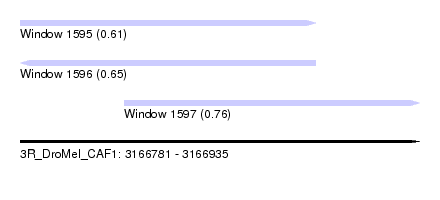
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,166,781 – 3,166,935 |
| Length | 154 |
| Max. P | 0.760994 |

| Location | 3,166,781 – 3,166,895 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -30.22 |
| Energy contribution | -30.67 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.610439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3166781 114 + 27905053 GUUACAGUUUCUAAUUUAUGACAAAUGCACAUGCCCGUCUUGAAGUUUUUGGCCUGGCCAGAAACUCGGCUCAAAU------CUGGACUGGACCAAGCCCAGGUCCAGCCCACCUCCAUU ...................(((....((....))..)))((((((((((((((...))))))))))....))))..------.(((.(((((((.......))))))).)))........ ( -30.80) >DroSec_CAF1 14862 114 + 1 GUUACAGUUUCUAAUUUAUGACAAAUGCACAUGCCCGUCUUGAAGUUUUUGGCCUGACCAGAAACUCGGCUGAAAU------CUGGACUGGACCAAGCCCAGGUCCAGCCCACCUCCAUU ....(((((.....((((.(((....((....))..))).))))((((((((.....))))))))..)))))....------.(((.(((((((.......))))))).)))........ ( -32.30) >DroEre_CAF1 6503 120 + 1 GUUACAGUUUCUAAUUUAUGACAAAUGCACAUGCCCGUCUUGAAGUUUUUGGCCUGGCCAGAAACUGAGCUGAAAUCCGAAACUGGACUGGACCGAGCUCAGGUCCCGCCCACCUCCAUU (((.(((((((....(((.(((....((....))..))).)))((((((((((...))))))))))............)))))))))).(((((.......))))).............. ( -32.70) >consensus GUUACAGUUUCUAAUUUAUGACAAAUGCACAUGCCCGUCUUGAAGUUUUUGGCCUGGCCAGAAACUCGGCUGAAAU______CUGGACUGGACCAAGCCCAGGUCCAGCCCACCUCCAUU ....(((((.....((((.(((....((....))..))).))))((((((((.....))))))))..)))))...........(((.(((((((.......))))))).)))........ (-30.22 = -30.67 + 0.45)

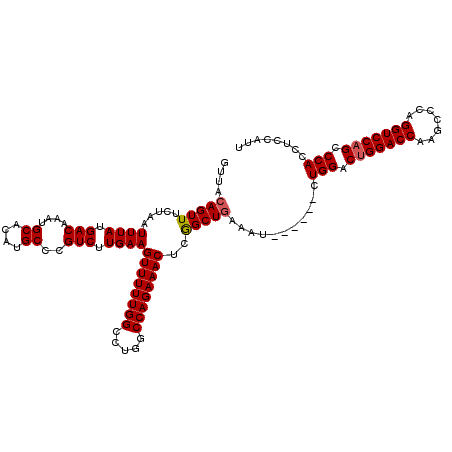
| Location | 3,166,781 – 3,166,895 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -34.09 |
| Energy contribution | -34.20 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.646087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3166781 114 - 27905053 AAUGGAGGUGGGCUGGACCUGGGCUUGGUCCAGUCCAG------AUUUGAGCCGAGUUUCUGGCCAGGCCAAAAACUUCAAGACGGGCAUGUGCAUUUGUCAUAAAUUAGAAACUGUAAC ..(((((.(((((((((((.......))))))))))).------.((((.(((..((.....))..)))))))..))))).((((.((....))...))))................... ( -39.40) >DroSec_CAF1 14862 114 - 1 AAUGGAGGUGGGCUGGACCUGGGCUUGGUCCAGUCCAG------AUUUCAGCCGAGUUUCUGGUCAGGCCAAAAACUUCAAGACGGGCAUGUGCAUUUGUCAUAAAUUAGAAACUGUAAC ..(((((.(((((((((((.......))))))))))).------.)))))....(((((((((.....))...........((((.((....))...)))).......)))))))..... ( -38.50) >DroEre_CAF1 6503 120 - 1 AAUGGAGGUGGGCGGGACCUGAGCUCGGUCCAGUCCAGUUUCGGAUUUCAGCUCAGUUUCUGGCCAGGCCAAAAACUUCAAGACGGGCAUGUGCAUUUGUCAUAAAUUAGAAACUGUAAC ..(((((.(((((.(((((.......))))).))))).)))))..........(((((((((((...)))...........((((.((....))...)))).......)))))))).... ( -38.60) >consensus AAUGGAGGUGGGCUGGACCUGGGCUUGGUCCAGUCCAG______AUUUCAGCCGAGUUUCUGGCCAGGCCAAAAACUUCAAGACGGGCAUGUGCAUUUGUCAUAAAUUAGAAACUGUAAC ........(((((((((((.......))))))))))).................((((((((((...)))...........((((.((....))...)))).......)))))))..... (-34.09 = -34.20 + 0.11)

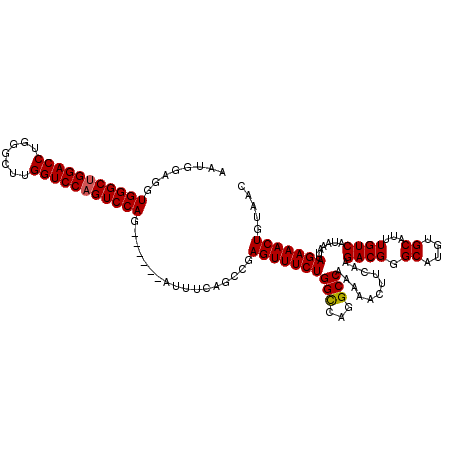
| Location | 3,166,821 – 3,166,935 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.66 |
| Mean single sequence MFE | -40.97 |
| Consensus MFE | -38.15 |
| Energy contribution | -38.27 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.760994 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3166821 114 + 27905053 UGAAGUUUUUGGCCUGGCCAGAAACUCGGCUCAAAU------CUGGACUGGACCAAGCCCAGGUCCAGCCCACCUCCAUUUGUGUAUCGUCACUCGAGGAAACUUAUUGGGCGAUGGGUU .((.(((((((((...)))))))))))(((((....------.(((.(((((((.......))))))).))).............((((((....(((....)))....))))))))))) ( -42.60) >DroSec_CAF1 14902 114 + 1 UGAAGUUUUUGGCCUGACCAGAAACUCGGCUGAAAU------CUGGACUGGACCAAGCCCAGGUCCAGCCCACCUCCAUUUGUGUAUCGUCACUCGAGGAAACUUAUUGGGCGAUGGGUU .((.((((((((.....))))))))))(((((..((------((((.((......)).)))))).)))))(((........)))(((((((....(((....)))....))))))).... ( -39.30) >DroEre_CAF1 6543 120 + 1 UGAAGUUUUUGGCCUGGCCAGAAACUGAGCUGAAAUCCGAAACUGGACUGGACCGAGCUCAGGUCCCGCCCACCUCCAUUUGUGUAUCGUCACUCGAGGAAACUUAUUGGGCGAUGGGUU ...((((((((((...)))))))))).......((((((..........(((((.......)))))((((((.........(((......)))..(((....)))..)))))).)))))) ( -41.00) >consensus UGAAGUUUUUGGCCUGGCCAGAAACUCGGCUGAAAU______CUGGACUGGACCAAGCCCAGGUCCAGCCCACCUCCAUUUGUGUAUCGUCACUCGAGGAAACUUAUUGGGCGAUGGGUU ...(((((((((.....))))))))).((((............(((.(((((((.......))))))).)))............(((((((....(((....)))....))))))))))) (-38.15 = -38.27 + 0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:39 2006