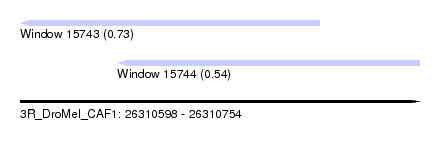
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,310,598 – 26,310,754 |
| Length | 156 |
| Max. P | 0.730174 |

| Location | 26,310,598 – 26,310,715 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -30.91 |
| Consensus MFE | -21.10 |
| Energy contribution | -21.73 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.730174 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26310598 117 - 27905053 CUCAGC-CACUGUGCGUUCUUUUGCAUUCCCGGCUUUGGAAUUUCAUUCACUGGCAUUGCAACCUACGCAUAUCUCUAUAUAAAUAAGCCUCACUAAUGAUUCUGCUGGCUUGG--GCAA ....((-(..(((((((....(((((...((((...((((......))))))))...)))))...)))))))............((((((.................)))))))--)).. ( -30.63) >DroSec_CAF1 35095 119 - 1 CUAAGC-CACUGUGCGUUCUUUUGCAUUCCCGGCUUUGGAAUUCCAUUCACCGGCAUUGCAACCUACGCAUAUCUCUAUAUAAAUAAGCCUCACUAAUGAUUCUGCUGGCUUGGGGGCAA ((((((-(..(((((((....(((((...((((...(((....)))....))))...)))))...)))))))..............(((.(((....)))....))))))))))...... ( -36.20) >DroSim_CAF1 36009 118 - 1 CGCAGCCCACUGUGCGUUCUUUUGCAUUCCCGGCUUUUGAAUUCCAUUCACUGGCAUUGCAACCUACGCAUAUCUCUAUAUAAAUAAGCCUCACUAAUGAUUCUGCUGGCUUGG--GCAA ....((((..(((((((....(((((...((((....(((((...)))))))))...)))))...))))))).............(((((.................)))))))--)).. ( -32.83) >DroYak_CAF1 36277 108 - 1 ----------UGUGCGUUCUGUUUCAUUCACGGCUAUGGAAUUCCAUUCACUGGCAUUGCAACCUACGCAUAUCUGCAUAUAAAUAAGCCCUACUAAUGAUUCUGCUGCCUUGG--GCAA ----------(((((((...(((.((....(((..((((....))))...)))....)).)))..)))))))...............((((.....................))--)).. ( -24.00) >consensus CUCAGC_CACUGUGCGUUCUUUUGCAUUCCCGGCUUUGGAAUUCCAUUCACUGGCAUUGCAACCUACGCAUAUCUCUAUAUAAAUAAGCCUCACUAAUGAUUCUGCUGGCUUGG__GCAA ..........(((((((....(((((...((((...(((....)))....))))...)))))...)))))))............((((((.................))))))....... (-21.10 = -21.73 + 0.63)

| Location | 26,310,636 – 26,310,754 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.41 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -20.48 |
| Energy contribution | -21.10 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26310636 118 - 27905053 -GCCACAUUUGUAAGGUUGCCUUGGUUUUUGUAUAUGUUUCUCAGC-CACUGUGCGUUCUUUUGCAUUCCCGGCUUUGGAAUUUCAUUCACUGGCAUUGCAACCUACGCAUAUCUCUAUA -.........(((.((((((....(((..((.....(((((..(((-(...(((((......)))))....))))..)))))......))..)))...)))))))))............. ( -28.90) >DroSec_CAF1 35135 118 - 1 -GCCACAUUUGUAAGGUUGCCUUGGAUUUUGUAUAUGUUUCUAAGC-CACUGUGCGUUCUUUUGCAUUCCCGGCUUUGGAAUUCCAUUCACCGGCAUUGCAACCUACGCAUAUCUCUAUA -.............(((.((.(((((.............)))))))-.)))((((((....(((((...((((...(((....)))....))))...)))))...))))))......... ( -29.32) >DroSim_CAF1 36047 119 - 1 -GCCACAUUUGUAAGGUUGCCUUGGAUUUUGUACAUGUUUCGCAGCCCACUGUGCGUUCUUUUGCAUUCCCGGCUUUUGAAUUCCAUUCACUGGCAUUGCAACCUACGCAUAUCUCUAUA -.........(((.((((((...(((...((((...(...((((........))))..)...)))).)))..(((..(((((...)))))..)))...)))))))))............. ( -26.50) >DroYak_CAF1 36315 107 - 1 UACGACAUUUGUAAGGAUGCCUUGGAGUUUGUUUAUA-------------UGUGCGUUCUGUUUCAUUCACGGCUAUGGAAUUCCAUUCACUGGCAUUGCAACCUACGCAUAUCUGCAUA ...((((.((.(((((...))))).))..))))...(-------------(((((((...(((.((....(((..((((....))))...)))....)).)))..))))))))....... ( -23.60) >consensus _GCCACAUUUGUAAGGUUGCCUUGGAUUUUGUAUAUGUUUCUCAGC_CACUGUGCGUUCUUUUGCAUUCCCGGCUUUGGAAUUCCAUUCACUGGCAUUGCAACCUACGCAUAUCUCUAUA ....(((..(.(((((...))))).)...)))..................(((((((....(((((...((((...(((....)))....))))...)))))...)))))))........ (-20.48 = -21.10 + 0.62)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:43:49 2006