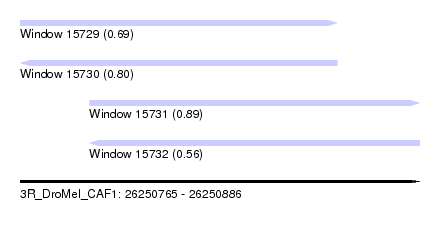
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 26,250,765 – 26,250,886 |
| Length | 121 |
| Max. P | 0.893992 |

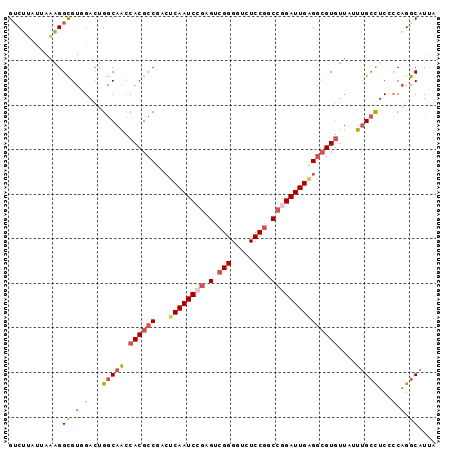
| Location | 26,250,765 – 26,250,861 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -38.80 |
| Consensus MFE | -25.43 |
| Energy contribution | -26.47 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692434 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26250765 96 + 27905053 GUCUUAUUAAGGGCGUUGACUGGCAACCACGCCUACUCAAUCCGAGUCGGGGUCUCCGUCCGGAUUGAGGCGUGUUAUUUGCCUCACCAGGCAUUA (((((....)))))((((.....))))((((((...((((((((.(.(((.....))).))))))))))))))).....(((((....)))))... ( -37.70) >DroSec_CAF1 940 96 + 1 GUCUUAUUAAAGGCGUGGACUGGCAACCACGCCGACUCAAUCCGAGUCGGGGUCUCCGGCCGGAUUGAGGCGUGUUAUUUGCCUCCCCAGGCAUUA ............((.(((...(((((.((((((...((((((((.(((((.....)))))))))))))))))))....)))))...))).)).... ( -45.00) >DroSim_CAF1 933 96 + 1 GUCUUAUUAAAGGCGUAGACUGGCAACCACGCCGACUCAAUCCGAGUCGGGGUCUCCGGCCGGAUUGAGGCGUGUUAUUUGCCUCCCCAGGCAUUA (((((....)))))((.....(((((.((((((...((((((((.(((((.....)))))))))))))))))))....))))).......)).... ( -40.30) >DroEre_CAF1 1020 96 + 1 GGCGUAUCAAAGGCGUGGACUGGCAGCCACGCCUACUCAAUCCGAGUCGGGGUCUCCGGCCGGAUUGGGGCGUGUUAUUUGCCUCACCAGGCAUCA .((((.....((((((((........)))))))).(((((((((.(((((.....))))))))))))))))))......(((((....)))))... ( -47.10) >DroYak_CAF1 942 96 + 1 GUCUGAUCAAAGGCGUGGACUGGCAACCACGCCUACUCAAUCCGAGUCGGGGUCUCCGUCCAGAUUGAGGCGUGUUAAUUGCCUCCCCAGGCAUUU (((((.....((((((((........))))))))..((((((.(.(.(((.....))).)).))))))((((.......))))....))))).... ( -36.20) >DroAna_CAF1 997 96 + 1 GUCGCAAAAGUGGUGUAGAGUGACAAGCACGGCGGGCCAAUCCAAGACGGGGUCUCCUCCCCGAUUGUGGAGUAGAAAUAGAUUCCUCUGACAUUU (((((((...((((.(...(((.....)))...).))))........(((((......))))).))))(((((........)))))...))).... ( -26.50) >consensus GUCUUAUUAAAGGCGUGGACUGGCAACCACGCCGACUCAAUCCGAGUCGGGGUCUCCGGCCGGAUUGAGGCGUGUUAUUUGCCUCCCCAGGCAUUA ............((.(.(...(((((.((((((...((((((((.(.(((.....))).)))))))))))))))....)))))...).).)).... (-25.43 = -26.47 + 1.03)


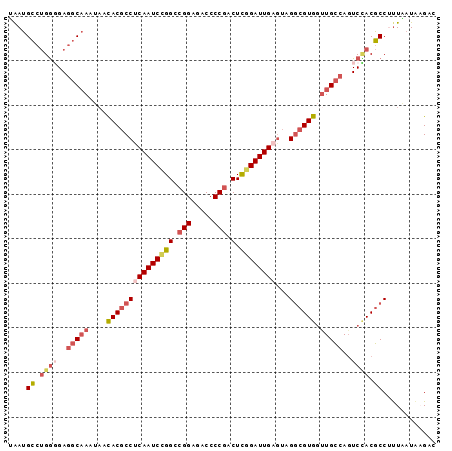
| Location | 26,250,765 – 26,250,861 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -35.63 |
| Consensus MFE | -28.33 |
| Energy contribution | -30.00 |
| Covariance contribution | 1.67 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.802097 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26250765 96 - 27905053 UAAUGCCUGGUGAGGCAAAUAACACGCCUCAAUCCGGACGGAGACCCCGACUCGGAUUGAGUAGGCGUGGUUGCCAGUCAACGCCCUUAAUAAGAC ........((((.(((((....(((((((((((((((.(((.....))).).))))))))...)))))).)))))......))))........... ( -35.80) >DroSec_CAF1 940 96 - 1 UAAUGCCUGGGGAGGCAAAUAACACGCCUCAAUCCGGCCGGAGACCCCGACUCGGAUUGAGUCGGCGUGGUUGCCAGUCCACGCCUUUAAUAAGAC ....((.((((..(((((....(((((((((((((((.(((.....))).).))))))))...)))))).)))))..)))).))............ ( -41.60) >DroSim_CAF1 933 96 - 1 UAAUGCCUGGGGAGGCAAAUAACACGCCUCAAUCCGGCCGGAGACCCCGACUCGGAUUGAGUCGGCGUGGUUGCCAGUCUACGCCUUUAAUAAGAC ....((.((((..(((((....(((((((((((((((.(((.....))).).))))))))...)))))).)))))..)))).))............ ( -39.30) >DroEre_CAF1 1020 96 - 1 UGAUGCCUGGUGAGGCAAAUAACACGCCCCAAUCCGGCCGGAGACCCCGACUCGGAUUGAGUAGGCGUGGCUGCCAGUCCACGCCUUUGAUACGCC ...(((((....)))))........((..((((((((.(((.....))).).)))))))...(((((((((.....).)))))))).......)). ( -36.20) >DroYak_CAF1 942 96 - 1 AAAUGCCUGGGGAGGCAAUUAACACGCCUCAAUCUGGACGGAGACCCCGACUCGGAUUGAGUAGGCGUGGUUGCCAGUCCACGCCUUUGAUCAGAC (((.((.((((..((((((...(((((((((((((((.(((.....))).).))))))))...))))))))))))..)))).)).)))........ ( -38.60) >DroAna_CAF1 997 96 - 1 AAAUGUCAGAGGAAUCUAUUUCUACUCCACAAUCGGGGAGGAGACCCCGUCUUGGAUUGGCCCGCCGUGCUUGUCACUCUACACCACUUUUGCGAC ....(((((.((((.....))))..((((....(((((......)))))...))))))))).(((.(((..(((......))).)))....))).. ( -22.30) >consensus UAAUGCCUGGGGAGGCAAAUAACACGCCUCAAUCCGGCCGGAGACCCCGACUCGGAUUGAGUAGGCGUGGUUGCCAGUCCACGCCUUUAAUAAGAC ....((.((((..(((((....(((((((((((((((.(((.....))).).))))))))...)))))).)))))..)))).))............ (-28.33 = -30.00 + 1.67)


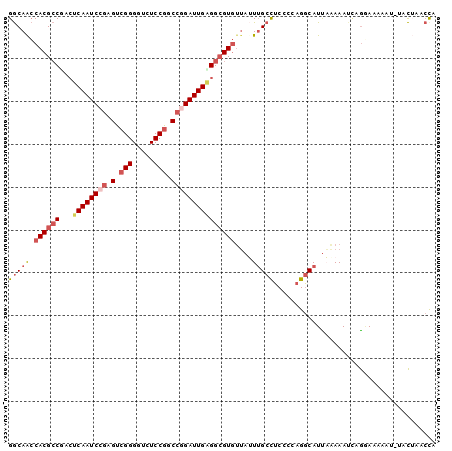
| Location | 26,250,786 – 26,250,886 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -32.39 |
| Consensus MFE | -23.40 |
| Energy contribution | -24.68 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26250786 100 + 27905053 GGCAACCACGCCUACUCAAUCCGAGUCGGGGUCUCCGUCCGGAUUGAGGCGUGUUAUUUGCCUCACCAGGCAUUAAAAAUUAGAAAAAACGUACUAACCA ((....((((((...((((((((.(.(((.....))).))))))))))))))).....(((((....))))).........................)). ( -33.70) >DroSec_CAF1 961 99 + 1 GGCAACCACGCCGACUCAAUCCGAGUCGGGGUCUCCGGCCGGAUUGAGGCGUGUUAUUUGCCUCCCCAGGCAUUAAAAAUCAGGAAAAAU-UACUAACCA ((....((((((...((((((((.(((((.....))))))))))))))))))).....(((((....)))))..................-......)). ( -37.00) >DroSim_CAF1 954 99 + 1 GGCAACCACGCCGACUCAAUCCGAGUCGGGGUCUCCGGCCGGAUUGAGGCGUGUUAUUUGCCUCCCCAGGCAUUAAAAAUCAGGAAAAAU-UACUAACCA ((....((((((...((((((((.(((((.....))))))))))))))))))).....(((((....)))))..................-......)). ( -37.00) >DroEre_CAF1 1041 99 + 1 GGCAGCCACGCCUACUCAAUCCGAGUCGGGGUCUCCGGCCGGAUUGGGGCGUGUUAUUUGCCUCACCAGGCAUCACAAAACAGGAAAAAU-UACGAUCCA (((((.(((((((...(((((((.(((((.....)))))))))))))))))))....)))))......((.(((................-...))))). ( -37.01) >DroYak_CAF1 963 99 + 1 GGCAACCACGCCUACUCAAUCCGAGUCGGGGUCUCCGUCCAGAUUGAGGCGUGUUAAUUGCCUCCCCAGGCAUUUAAAAUAAGAAAGAAG-UACGAUCUA .((...((((((...((((((.(.(.(((.....))).)).))))))))))))((((.(((((....))))).))))............)-)........ ( -28.80) >DroAna_CAF1 1018 97 + 1 GACAAGCACGGCGGGCCAAUCCAAGACGGGGUCUCCUCCCCGAUUGUGGAGUAGAAAUAGAUUCCUCUGACAUUUUAAAUCCAUAAAAAUAUUUU---CA .........((....)).........(((((......))))).(((((((.(((((.((((....))))...)))))..))))))).........---.. ( -20.80) >consensus GGCAACCACGCCGACUCAAUCCGAGUCGGGGUCUCCGGCCGGAUUGAGGCGUGUUAUUUGCCUCCCCAGGCAUUAAAAAUCAGGAAAAAU_UACUAACCA (((((.((((((...((((((((.(.(((.....))).)))))))))))))))....)))))...................................... (-23.40 = -24.68 + 1.28)


| Location | 26,250,786 – 26,250,886 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -31.91 |
| Consensus MFE | -25.05 |
| Energy contribution | -26.33 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563442 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 26250786 100 - 27905053 UGGUUAGUACGUUUUUUCUAAUUUUUAAUGCCUGGUGAGGCAAAUAACACGCCUCAAUCCGGACGGAGACCCCGACUCGGAUUGAGUAGGCGUGGUUGCC .(((....((..................(((((....))))).....(((((((((((((((.(((.....))).).))))))))...)))))))).))) ( -34.80) >DroSec_CAF1 961 99 - 1 UGGUUAGUA-AUUUUUCCUGAUUUUUAAUGCCUGGGGAGGCAAAUAACACGCCUCAAUCCGGCCGGAGACCCCGACUCGGAUUGAGUCGGCGUGGUUGCC .........-....(((((..............)))))(((((....(((((((((((((((.(((.....))).).))))))))...)))))).))))) ( -35.64) >DroSim_CAF1 954 99 - 1 UGGUUAGUA-AUUUUUCCUGAUUUUUAAUGCCUGGGGAGGCAAAUAACACGCCUCAAUCCGGCCGGAGACCCCGACUCGGAUUGAGUCGGCGUGGUUGCC .........-....(((((..............)))))(((((....(((((((((((((((.(((.....))).).))))))))...)))))).))))) ( -35.64) >DroEre_CAF1 1041 99 - 1 UGGAUCGUA-AUUUUUCCUGUUUUGUGAUGCCUGGUGAGGCAAAUAACACGCCCCAAUCCGGCCGGAGACCCCGACUCGGAUUGAGUAGGCGUGGCUGCC .(((.....-.....)))....((((..(((((....))))).))))(((((((((((((((.(((.....))).).))))))).)..))))))...... ( -33.30) >DroYak_CAF1 963 99 - 1 UAGAUCGUA-CUUCUUUCUUAUUUUAAAUGCCUGGGGAGGCAAUUAACACGCCUCAAUCUGGACGGAGACCCCGACUCGGAUUGAGUAGGCGUGGUUGCC ......(((-.............((((.(((((....))))).))))(((((((((((((((.(((.....))).).))))))))...))))))..))). ( -32.00) >DroAna_CAF1 1018 97 - 1 UG---AAAAUAUUUUUAUGGAUUUAAAAUGUCAGAGGAAUCUAUUUCUACUCCACAAUCGGGGAGGAGACCCCGUCUUGGAUUGGCCCGCCGUGCUUGUC ..---..........(((((.........(((((.((((.....))))..((((....(((((......)))))...)))))))))...)))))...... ( -20.10) >consensus UGGUUAGUA_AUUUUUCCUGAUUUUUAAUGCCUGGGGAGGCAAAUAACACGCCUCAAUCCGGCCGGAGACCCCGACUCGGAUUGAGUAGGCGUGGUUGCC ............................(((((....))))).....(((((((((((((((.(((.....))).).))))))))...))))))...... (-25.05 = -26.33 + 1.28)

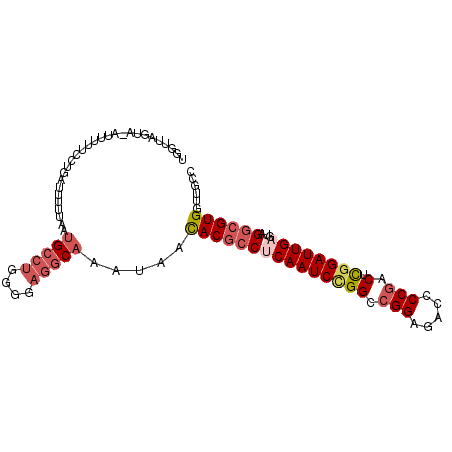
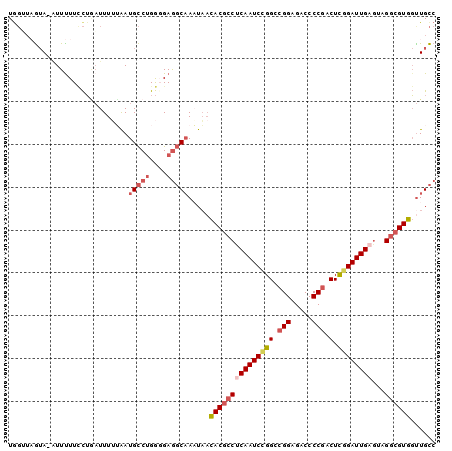
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 13:43:38 2006