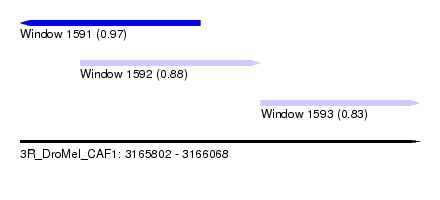
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,165,802 – 3,166,068 |
| Length | 266 |
| Max. P | 0.970520 |

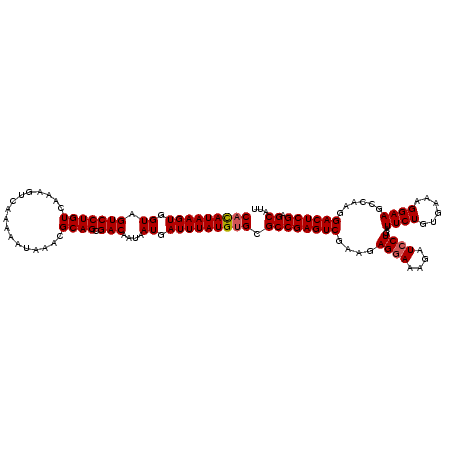
| Location | 3,165,802 – 3,165,922 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -29.11 |
| Energy contribution | -29.35 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3165802 120 - 27905053 CACAUAAGUGGUAGUCCUGUCAAAGUCAAAAAUAAACGCAGCGACAAUAAUGAUUUAUGUGCGCCGAGUCGAAGAGAAAAGAUCCUGAUUCUGUGAAAGGAAGCCAAGGACUCGAGCAUU (((((((((.((.(((((((.................)))).)))....)).))))))))).((((((((.....)).....((((..((((......))))....)))))))).))... ( -28.23) >DroSec_CAF1 13884 120 - 1 CACAUAAGUGGUAGUCCUGUCAAAGUCAAAAAUAAACGCAGCGACAAUAAUGAUUUAUGUGCGCCGAGUCGAAGAGGAAAGAUCCUGAUUCUGUGAAAGGAAGCCAAGGACUCGAGCAUU (((((((((.((.(((((((.................)))).)))....)).))))))))).((((((((....((((....))))..((((......))))......)))))).))... ( -31.73) >DroSim_CAF1 16602 120 - 1 CACAUAAGUGGUAGUCCUGUCAAAGUCAAAAAUAAACGCAGCGACAAUAAUGAUUUAUGUGCGCCGAGUCGAAGAGGAAAGAUCCUGAUUCUGUAAAAGGAAGCCAAGGACUCGAGCAGU (((((((((.((.(((((((.................)))).)))....)).))))))))).((((((((....((((....))))..((((......))))......)))))).))... ( -31.73) >DroEre_CAF1 5557 120 - 1 CACAUAAGUGGUAGUCCUGUCAAAGUCAAAAAUAAACGCAGCGACAAUAAUGAUUUAUGGGCGCCGAGUCGAAAAGGAAAGCUCCUGAUUCUGCGAAAGGAAGCCCAGGACUCGAGCAUU ..(((((((.((.(((((((.................)))).)))....)).)))))))...(((((((((((.((((....))))..)))..(....).........)))))).))... ( -30.43) >DroYak_CAF1 15133 120 - 1 CAUAUAAGUGGUAGUCCUGUCAAAGUCAAAAAUAAACGCAGCGACAAUAAUGAUUUAUGUGCGCCGAGUCGAAAAGGAAAGGUCCUGAUUCUGUGGAAGGAAGCCAAGGACUCGAGCAUU (((((((((.((.(((((((.................)))).)))....)).))))))))).(((((((((((.((((....))))..)))..(((.......)))..)))))).))... ( -31.43) >consensus CACAUAAGUGGUAGUCCUGUCAAAGUCAAAAAUAAACGCAGCGACAAUAAUGAUUUAUGUGCGCCGAGUCGAAGAGGAAAGAUCCUGAUUCUGUGAAAGGAAGCCAAGGACUCGAGCAUU (((((((((.((.(((((((.................)))).)))....)).))))))))).((((((((....((((....))))..((((......))))......)))))).))... (-29.11 = -29.35 + 0.24)

| Location | 3,165,842 – 3,165,962 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -27.94 |
| Energy contribution | -27.62 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3165842 120 + 27905053 UUUUCUCUUCGACUCGGCGCACAUAAAUCAUUAUUGUCGCUGCGUUUAUUUUUGACUUUGACAGGACUACCACUUAUGUGUUUAUGUCGGCGUUGAGAAGAAGAAGCUGAGAAGGACGAA ((((((((((..(((((((((((((((((((...(((((..(((........)).)..)))))((....))....))).))))))))..))))))))..)))))....)))))....... ( -31.70) >DroSec_CAF1 13924 112 + 1 UUUCCUCUUCGACUCGGCGCACAUAAAUCAUUAUUGUCGCUGCGUUUAUUUUUGACUUUGACAGGACUACCACUUAUGUGUUUAUGUCGGCGUUGAGAAGGAGAAUCUGAGC-------- ((((((..((((((((((((((((((........(((((..(((........)).)..)))))((....))..))))))))....))))).)))))..))))))........-------- ( -31.40) >DroSim_CAF1 16642 112 + 1 UUUCCUCUUCGACUCGGCGCACAUAAAUCAUUAUUGUCGCUGCGUUUAUUUUUGACUUUGACAGGACUACCACUUAUGUGUUUAUGUCGGCGUUGAGAAGGAGAAUCUGAGC-------- ((((((..((((((((((((((((((........(((((..(((........)).)..)))))((....))..))))))))....))))).)))))..))))))........-------- ( -31.40) >DroEre_CAF1 5597 112 + 1 UUUCCUUUUCGACUCGGCGCCCAUAAAUCAUUAUUGUCGCUGCGUUUAUUUUUGACUUUGACAGGACUACCACUUAUGUGUUUAUGUUGGCGUUGAGCAGGAGAAACCGAGC-------- .....(((((..(((((((((((((((((((...(((((..(((........)).)..)))))((....))....))).)))))))..)))))))))..)))))........-------- ( -29.40) >DroYak_CAF1 15173 112 + 1 UUUCCUUUUCGACUCGGCGCACAUAAAUCAUUAUUGUCGCUGCGUUUAUUUUUGACUUUGACAGGACUACCACUUAUAUGGUUAUGUUGGCGUUGAGAAGGAGGAACUGAGC-------- (((((((((((((.(((((.(((...........)))))))).))).......(((.(..(((.....((((......))))..)))..).)))))))))))))........-------- ( -30.60) >consensus UUUCCUCUUCGACUCGGCGCACAUAAAUCAUUAUUGUCGCUGCGUUUAUUUUUGACUUUGACAGGACUACCACUUAUGUGUUUAUGUCGGCGUUGAGAAGGAGAAACUGAGC________ .....(((((..(((((((((((((((.(((...(((((..(((........)).)..)))))((....))......))))))))))..))))))))..)))))................ (-27.94 = -27.62 + -0.32)

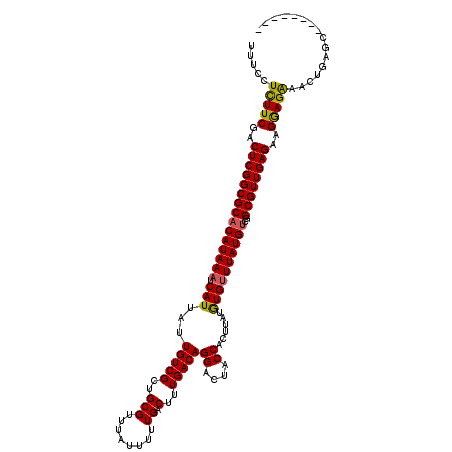
| Location | 3,165,962 – 3,166,068 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.69 |
| Mean single sequence MFE | -28.04 |
| Consensus MFE | -15.28 |
| Energy contribution | -16.56 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.831101 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3165962 106 + 27905053 GAAGGAAAAUGGAAUGUGACUCCCAAGGAGAAUGAAGAACAGUGGUAGACGUCUACCAAUUCAUUCUCAUUC---------AUAUUU-CUA-CUUCUUUAAACAUUUCAA---UAGAGAA .((((((...(((((((((((....))(((((((((......((((((....)))))).)))))))))..))---------))))))-)..-.)))))).....((((..---..)))). ( -31.70) >DroSec_CAF1 14036 114 + 1 --AGGAAAAUGGGAUGGGACUCCCAAGAAGAAUGAAGAAUAGUGGUAGACGUCUACCAAUUCAUUCUUAUUCUCAGCAGUAAUAUUUGUUA-CUUCUUUAAACAUUUCAU---UAGAGAA --.......(((((((((...))))..(((((((((......((((((....)))))).)))))))))..)))))..((((((....))))-))(((((((........)---)))))). ( -31.50) >DroSim_CAF1 16754 114 + 1 --AGGAAAAUGGGAUGGGACUCCCAAGGAGAAUGAAGAAUAGUGGUAGACGUCUACCAAUUCAUUCAUAUUCUCAGCAGUAAUAUUUGUUA-CUUCUUUAAACAUUUCAA---UAGAGAA --..((((.(((((......)))))..(((((((((((((..((((((....)))))))))).)))))..))))((.((((((....))))-)).)).......))))..---....... ( -29.50) >DroEre_CAF1 5709 118 + 1 --AGGAAAAUGGGAUGGGACUCACAAACAGCAUGGGGAAUAGUAGUAGACGUCUACCAAAUGAUUCUCAUUCGCAGCAGUAAUAUUUUUCAAGUUUUUCAAGGGAUUGCGAUCCCUUUAA --.((((((((((((((((.(((((.......))..........((((....))))....))).))))))))((....))..)))))))).........(((((((....)))))))... ( -25.90) >DroYak_CAF1 15285 115 + 1 --AGGAAAAUGGUAUGGGACUCCCAAGGAGCAUGAAGAAUAGUGGCAGACGUCUACCAAGUGAUUCUCAUUCGCAGCAGUACAAUUUGUUAACUUCUUGAAACAUUUGAA---CAGAAAA --.(..(((((....((....))(((((((.((((.((((..(((.((....)).)))....))))))))....(((((......)))))..)))))))...)))))...---)...... ( -21.60) >consensus __AGGAAAAUGGGAUGGGACUCCCAAGGAGAAUGAAGAAUAGUGGUAGACGUCUACCAAUUCAUUCUCAUUCGCAGCAGUAAUAUUUGUUA_CUUCUUUAAACAUUUCAA___UAGAGAA ..(((((..(((((......)))))....((((((.((((..((((((....))))))....)))))))))).....................)))))...................... (-15.28 = -16.56 + 1.28)

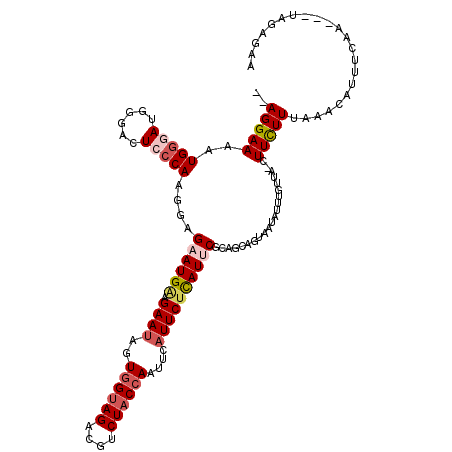
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:35 2006