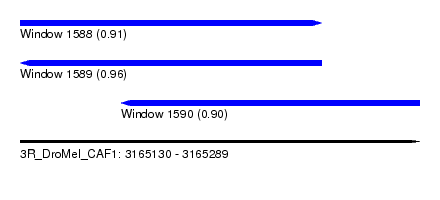
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 3,165,130 – 3,165,289 |
| Length | 159 |
| Max. P | 0.959908 |

| Location | 3,165,130 – 3,165,250 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -43.18 |
| Consensus MFE | -36.70 |
| Energy contribution | -37.95 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.85 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3165130 120 + 27905053 GCACAGCCUCGGCAAGGAAUGAGGUAGUGCUGGGUGGUCGGGGUCGAUUGGGUCUGCUGUGCAAAUUUAAAUUUGGUAAUGGUCCUGGCACUUUUUGCCACGGACUGUAAAGCAAAUAAG ((((((((((((((..(.......)..)))))))......((..(.....)..)))))))))........(((((.(.((((((((((((.....))))).)))))))..).)))))... ( -40.90) >DroSec_CAF1 13193 120 + 1 GCACAGCCUCGGCAAGGAAUGAGGUAGUGCUGGGGGGUCGGGGUCGAUUGGGUCUGCUGUGCAAAUUUAAAUUUGGUAAUGGUCCUGGCACUUUUUGCCAGGGACUGUUAAGCAAAUAAG ((((((((((((((..(.......)..)))))))((..(.((.....)).)..)))))))))........(((((.((((((((((((((.....))))).)))))))))..)))))... ( -45.90) >DroSim_CAF1 15432 120 + 1 GCACAGCCUCGGAAAGGAAUGAGGUAGUGCUGGGUGGUCGGGGUCGAUUGGGUCUGCUGUGCAAAUUUAAAUUUGGUAAUGGUCCUGGCACUUUUUGCCAGGGACUGUUAAGCAAAUAAG ((((.((((((........)))))).))))((.(..(.((((.((....)).)))))..).)).......(((((.((((((((((((((.....))))).)))))))))..)))))... ( -44.40) >DroEre_CAF1 4891 120 + 1 GCACAGCCUCGGUAAGGAAUGAGGUAGUGCUGAGUGGUCGGGGUCGAUUGGGUCUGCUGUGCAAAUUUAAAUUCGGUAAUGGUCCUGCCACUUUUUGCCAGAGACUGCUAGGCAAAUAAG ((((.((((((........)))))).)))).((((((.((((..((((...)))(((((..............)))))..)..))))))))))((((((((......)).)))))).... ( -41.54) >consensus GCACAGCCUCGGCAAGGAAUGAGGUAGUGCUGGGUGGUCGGGGUCGAUUGGGUCUGCUGUGCAAAUUUAAAUUUGGUAAUGGUCCUGGCACUUUUUGCCAGGGACUGUUAAGCAAAUAAG ((((.((((((........)))))).))))((.(..(.((((.((....)).)))))..).)).......(((((.((((((((((((((.....))))).)))))))))..)))))... (-36.70 = -37.95 + 1.25)

| Location | 3,165,130 – 3,165,250 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.57 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3165130 120 - 27905053 CUUAUUUGCUUUACAGUCCGUGGCAAAAAGUGCCAGGACCAUUACCAAAUUUAAAUUUGCACAGCAGACCCAAUCGACCCCGACCACCCAGCACUACCUCAUUCCUUGCCGAGGCUGUGC ....((((((.....((((.(((((.....)))))))))......(((((....)))))...)))))).....(((....))).......((((..((((..........))))..)))) ( -29.70) >DroSec_CAF1 13193 120 - 1 CUUAUUUGCUUAACAGUCCCUGGCAAAAAGUGCCAGGACCAUUACCAAAUUUAAAUUUGCACAGCAGACCCAAUCGACCCCGACCCCCCAGCACUACCUCAUUCCUUGCCGAGGCUGUGC ....((((((.....(..(((((((.....)))))))..).....(((((....)))))...)))))).....(((....))).......((((..((((..........))))..)))) ( -29.80) >DroSim_CAF1 15432 120 - 1 CUUAUUUGCUUAACAGUCCCUGGCAAAAAGUGCCAGGACCAUUACCAAAUUUAAAUUUGCACAGCAGACCCAAUCGACCCCGACCACCCAGCACUACCUCAUUCCUUUCCGAGGCUGUGC ....((((((.....(..(((((((.....)))))))..).....(((((....)))))...)))))).....(((....))).......((((..((((..........))))..)))) ( -29.80) >DroEre_CAF1 4891 120 - 1 CUUAUUUGCCUAGCAGUCUCUGGCAAAAAGUGGCAGGACCAUUACCGAAUUUAAAUUUGCACAGCAGACCCAAUCGACCCCGACCACUCAGCACUACCUCAUUCCUUACCGAGGCUGUGC ....((((((.((......)))))))).(((((..((........(((.......(((((...))))).....)))...))..)))))..((((..((((..........))))..)))) ( -26.70) >consensus CUUAUUUGCUUAACAGUCCCUGGCAAAAAGUGCCAGGACCAUUACCAAAUUUAAAUUUGCACAGCAGACCCAAUCGACCCCGACCACCCAGCACUACCUCAUUCCUUGCCGAGGCUGUGC ....(((((......((((.(((((.....)))))))))......(((((....)))))....))))).....(((....))).......((((..((((..........))))..)))) (-26.70 = -26.57 + -0.13)

| Location | 3,165,170 – 3,165,289 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.19 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -21.90 |
| Energy contribution | -22.65 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 3165170 119 - 27905053 AGGCAUAUUCCCAAGCCUUAGAAU-CAAAGAAGUAAAGCACUUAUUUGCUUUACAGUCCGUGGCAAAAAGUGCCAGGACCAUUACCAAAUUUAAAUUUGCACAGCAGACCCAAUCGACCC .(((..........)))(((((..-.......((((((((......)))))))).((((.(((((.....)))))))))..........))))).(((((...)))))............ ( -28.00) >DroSec_CAF1 13233 119 - 1 AGGCAUAUUACCAAGCCUUAGAAU-CCAAAAAGUUAAGCACUUAUUUGCUUAACAGUCCCUGGCAAAAAGUGCCAGGACCAUUACCAAAUUUAAAUUUGCACAGCAGACCCAAUCGACCC ((((..........))))......-.......((((((((......)))))))).((((((((((.....)))))))..................(((((...))))).......))).. ( -29.50) >DroSim_CAF1 15472 119 - 1 AGGCAUAUUACCAAGCCUUAGAAU-CCAAAAAGUAAAGCACUUAUUUGCUUAACAGUCCCUGGCAAAAAGUGCCAGGACCAUUACCAAAUUUAAAUUUGCACAGCAGACCCAAUCGACCC ((((..........))))......-.......((.(((((......))))).)).((((((((((.....)))))))..................(((((...))))).......))).. ( -25.60) >DroEre_CAF1 4931 117 - 1 AGGCAU--UC-CGAACCUUAGAAUUCCAGGAAGUAAAGCACUUAUUUGCCUAGCAGUCUCUGGCAAAAAGUGGCAGGACCAUUACCGAAUUUAAAUUUGCACAGCAGACCCAAUCGACCC (((((.--..-....(((.........)))((((.....))))...)))))....((((((((((((.(((((.....)))))............))))).))).))))........... ( -21.02) >consensus AGGCAUAUUACCAAGCCUUAGAAU_CCAAAAAGUAAAGCACUUAUUUGCUUAACAGUCCCUGGCAAAAAGUGCCAGGACCAUUACCAAAUUUAAAUUUGCACAGCAGACCCAAUCGACCC .(((..........)))(((((..........((((((((......)))))))).((((.(((((.....)))))))))..........))))).(((((...)))))............ (-21.90 = -22.65 + 0.75)

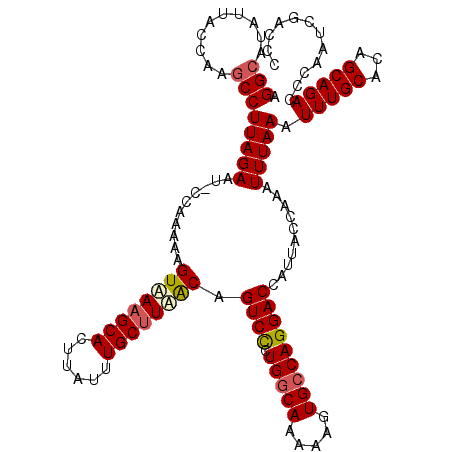
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:57:33 2006